Abstract
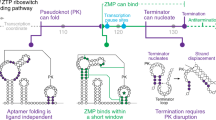
Riboswitches are genetic regulatory elements found in the 5′ untranslated region of messenger RNA that act in the absence of protein cofactors1,2. They are broadly distributed across bacteria and account for the regulation of more than 2% of all genes in Bacillus subtilis, underscoring their importance in the control of cellular metabolism3. The 5′ untranslated region of many mRNAs of genes involved in purine metabolism and transport contain a guanine-responsive riboswitch that directly binds guanine, hypoxanthine or xanthine to terminate transcription3,4. Here we report the crystal structure at 1.95 Å resolution of the purine-binding domain of the guanine riboswitch from the xpt–pbuX operon of B. subtilis bound to hypoxanthine, a prevalent metabolite in the bacterial purine salvage pathway. This structure reveals a complex RNA fold involving several phylogenetically conserved nucleotides that create a binding pocket that almost completely envelops the ligand. Hypoxanthine functions to stabilize this structure and to promote the formation of a downstream transcriptional terminator element, thereby providing a mechanism for directly repressing gene expression in response to an increase in intracellular concentrations of metabolite.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vitreschak, A. G., Rodionov, D. A., Mironov, A. A. & Gelfand, M. S. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 20, 44–50 (2004)
Mandal, M. & Breaker, R. R. Gene regulation by riboswitches. Nature Rev. Mol. Cell. Biol. 5, 451–463 (2004)
Mandal, M., Boese, B., Barrick, J. E., Winkler, W. C. & Breaker, R. R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113, 577–586 (2003)
Johansen, L. E., Nygaard, P., Lassen, C., Agerso, Y. & Saxild, H. H. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt–pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL). J. Bacteriol. 185, 5200–5209 (2003)
Ellington, A. D. & Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature 346, 818–822 (1990)
Gold, L., Polisky, B., Uhlenbeck, O. & Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 64, 763–797 (1995)
Silverman, S. K. Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA 9, 377–383 (2003)
Seetharaman, S., Zivarts, M., Sudarsan, N. & Breaker, R. R. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nature Biotechnol. 19, 336–341 (2001)
Zimmermann, G. R., Jenison, R. D., Wick, C. L., Simmorre, J.-P. & Pardi, A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nature Struct. Biol. 4, 644–649 (1997)
Fan, P., Suri, A. K., Fiala, R., Live, D. & Patel, D. J. Molecular recognition in the FMN–RNA aptamer complex. J. Mol. Biol. 258, 480–500 (1996)
Baugh, C., Grate, D. & Wilson, C. 2.8 Å crystal structure of the malachite green aptamer. J. Mol. Biol. 301, 117–128 (2000)
Koizumi, M., Soukup, G. A., Kerr, J. N. & Breaker, R. R. Allosteric selection of ribozymes that respond to the second messengers cGMP and cAMP. Nature Struct. Biol. 6, 1062–1071 (1999)
Leulliot, N. & Varani, G. Current topics in RNA–protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry 40, 7947–7956 (2001)
Williamson, J. R. Induced fit in RNA–protein recognition. Nature Struct. Biol. 7, 834–837 (2000)
Mandal, M. & Breaker, R. R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct. Mol. Biol. 11, 29–35 (2004)
Doherty, E. A., Batey, R. T., Masquida, B. & Doudna, J. A. A universal mode of helix packing in RNA. Nature Struct. Biol. 8, 339–343 (2001)
Nissen, P., Ippolito, J. A., Ban, N., Moore, P. B. & Steitz, T. A. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA 98, 4899–4903 (2001)
Cate, J. H. et al. RNA tertiary structure mediation by adenosine platforms. Science 273, 1696–1699 (1996)
Correll, C. C., Beneken, J., Plantinga, M. J., Lubbers, M. & Chan, Y. L. The common and the distinctive features of the bulged-G motif based on a 1.04 Å resolution RNA structure. Nucleic Acids Res. 31, 6806–6818 (2003)
Molinaro, M. & Tinoco, I. Jr Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 23, 3056–3063 (1995)
De la Pena, M., Gago, S. & Flores, R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 22, 5561–5570 (2003)
Khvorova, A., Lescoute, A., Westhof, E. & Jayasena, S. D. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nature Struct. Biol. 10, 708–712 (2003)
Choi, K. Y. & Zalkin, H. Structural characterization and corepressor binding of the Escherichia coli purine repressor. J. Bacteriol. 174, 6207–6214 (1992)
Kieft, J. S. & Batey, R. T. A general method for rapid and nondenaturing purification of RNAs. RNA 10, 988–995 (2004)
Pflugrath, J. W. The finer things in X-ray diffraction data collection. Acta Crystallogr. D 55, 1718–1725 (1999)
Terwilliger, T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J. Synchrotron Radiat. 11, 49–52 (2004)
Brunger, A. T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Schumacher, M. A., Choi, K. Y., Zalkin, H. & Brennan, R. G. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by α helices. Science 266, 763–770 (1994)
Acknowledgements
We thank S. Edwards for maintaining and managing the Biochemistry Division X-ray Crystallography facility; and T. Cech, A. Pardi, D. Wuttke, J. Kieft and R. Rambo for discussions and comments on the manuscript. This work was funded in part from a grant from the Research Corporation and the University of Colorado Butcher Biotechnology Initiative. S.D.G. was supported in part by a NIH predoctoral training grant.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Information
Additional description of methods, figures and references. (DOC 1854 kb)
Rights and permissions
About this article
Cite this article
Batey, R., Gilbert, S. & Montange, R. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 432, 411–415 (2004). https://doi.org/10.1038/nature03037
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03037
This article is cited by
-
Structure and mechanism of the methyltransferase ribozyme MTR1
Nature Chemical Biology (2022)
-
Targeting RNA structures with small molecules
Nature Reviews Drug Discovery (2022)
-
Characterization of Five Purine Riboswitches in Cellular and Cell-Free Expression Systems
Current Microbiology (2022)
-
Getting to the bottom of lncRNA mechanism: structure–function relationships
Mammalian Genome (2022)
-
The energy-spectrum of bicompatible sequences
Algorithms for Molecular Biology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.