Abstract
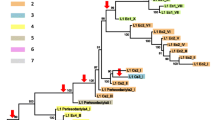
One of the uncertainties regarding the evolution of L1 elements is whether there are numerous progenitor genes. We present phylogenetic evidence from ORF1 sequences of slow loris (Nycticebus coucang) and galago (Galago crassicaudatus) that there were at least two distinct progenitors, active at the same time, in the ancestor of this family of prosimian primates. A maximum parsimony analysis that included representative L1s from human, rabbit, and rodents, along with the prosimian sequences, revealed that one of the galago L1s (Gc11) grouped very strongly with the slow loris sequences. The remaining galago elements formed their own unique and strongly supported clade. An analysis of replacement and silent site changes for each link of the most parsimonious tree indicated that during the descent of the Gc11 sequence approximately two times more synonymous than nonsynonymous substitutions had occurred, implying that the Gc11 founder was functional for some time after the split of galago and slow loris. Strong purifying selection was also evident on the galago branch of the tree. These data indicate that there were two distinct and contemporaneous L1 progenitors in the lorisoid ancestor, evolving under purifying selection, that were retained as functional L1s in the galago lineage (and presumably also in the slow loris). The prosimian ORF1 sequences could be further subdivided into subfamilies. ORF1 sequences from both the galago and slow loris have a premature termination codon near the 3′ end, not shared by the other mammalian sequences, that shortens the open reading frame by 288 bp. An analysis of synonymous and nonsynonymous substitutions for the 5′ and 3′ portions, that included intra- and inter-subfamily comparisons, as well as comparisons among the other mammalian sequences, suggested that this premature stop codon is a prosimian acquisition that has rendered the 3′ portion of ORF1 in these primates noncoding.
Similar content being viewed by others
References
Bailey WJ, Slightom JL, Goodman M (1992) Rejection of the “flying primate” hypothesis by phylogenetic evidence from the ε-globin gene. Science 256:86–89
Batzer MA, Deininger PL (1991) A human-specific subfamily of Alu sequences. Genomics 9:481–487
Besansky NJ (1990) A retrotransposable element from the mosquitoAnopheles gambiae. Mol Cell Biol 10:863–871
Britten RJ, Baron WF, Stout D, Davidson EH (1988) Sources and evolution of human Alu repeated sequences. Proc Natl Acad Sci USA 85:4770–4774
Burton FH, Loeb DD, Voliva CF, Martin SL, Edgell MH, Hutchison III CA (1986) Conservation throughout Mammalia and extensive protein-coding capacity of the highly repeated DNA long interspersed sequence one. J Mol Biol 187:291–304
Czelusniak J, Goodman M, Hewett-Emmett D, Weiss ML, Venta PJ, Tashian RE (1982) Phylogenetic origins and adaptive evolution of avian and mammalian haemoglobin. Nature 298:297–300
Czelusniak J, Goodman M, Moncrief ND, Kehoe SM (1990) Maximum parismony approach to construction of evolutionary trees from aligned homologous sequences. In: Doolittle RF (ed) Methods in enzymology. Academic Press, San Diego, vol 183 pp 601–615
D'Ambrosio E, Waitzkin SD, Witney FR, Salemme A, Furano AV (1986) Structure of the highly repeated, long interspersed DNA family (LINE or L1Rn) of the rat. Mol Cell Biol 6:411–424
Daniels GR, Fox GM, Loewensteiner D, Schmid CW, Deininger PL (1983) Species-specific homogeneity of the primate Alu family of repeated DNA sequences. Nucleic Acids Res 11:7579–7593
Demers GW, Matunis MJ, Hardison RC (1989) The L1 family of long interspersed repetitive DNA in rabbits: sequence, copy number, conserved open reading frames, and similarity to keratin. J Mol Evol 29:3–19
Di Nocera PP, Casari G (1987) Related polypeptides are encoded byDrosophila F elements, I factors, and mammalian L1 sequences. Proc Natl Acad Sci USA 84:5843–5847
Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian Jr HH (1991) Isolation of an active human transposable element. Science 254:1805–1808
Garrett JE, Knutzon DS, Carroll D (1989) Composite transposable elements in theXenopus laevis genome. Mol Cell Biol 9:3018–3027
Groves CP (1974) Taxonomy and phylogeny of prosimians. In: Martin RD, Doyle GA, Walker AC (eds) Prosimian biology. Gerald Duckworth, Liverpool, pp 449–473
Hardies SC, Martin SL, Voliva CF, Hutchison III CA, Edgell MH (1986) An analysis of replacement and synonymous changes in the rodent L1 repeat family. Mol Biol Evol 3:109–125
Hattori M, Hidaka S, Sakaki Y (1985) Sequence analysis of a KpnI family member near the 3′ end of human β-globin gene. Nucleic Acids Res 13:7813–7827
Hattori M, Kuhara S, Takenaka O, Sakaki Y (1986) L1 family of repetitive DNA sequences in primates may be derived from a sequence encoding a reverse transcriptase-related protein. Nature 321:625–628
Hendy MD, Penny D (1982) Branch and bound algorithms to determine minimal evolutionary trees. Math Biosci 59:277–290
Hohjoh H, Minakami R, Sakaki Y (1990) Selective cloning and sequence analysis of the human L1 (LINE-1) sequences which transposed in the relatively recent past. Nucleic Acids Res 18:4099–4104
Jukes TH, Cantor CR (1969) Evolution of protein molecules. In: Munro HN (ed) Mammalian protein metabolism. Academic Press, New York, vol 2 pp 21–123
Jurka J (1989) Subfamily structure and evolution of the human L1 family of repetitive sequences. J Mol Evol 29:496–503
Jurka J, Smith T (1988) A fundamental division in the Alu family of repeated sequences. Proc Natl Acad Sci USA 85:4775–4778
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge, England
Krane DE, Clark AG, Cheng J-F, Hardison RC (1991) Subfamily relationships and clustering of rabbit C repeats. Mol Biol Evol 8:1–30
Leeflang EP, Liu W-M, Hashimoto C, Choudary PV, Schmid CW (1992) Phylogenetic evidence for multiple Alu source genes. J Mol Evol 35:7–16
Leibold DA, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG (1990) Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci USA 87:6990–6994
Li W-H, Graur D (1991) Fundamentals of molecular evolution. Sinauer Associates, Sunderland, MA
Lloyd JA, Potter SS (1988) Distinct subfamilies of primate L1Gg retroposons, with some elements carrying tandem repeats in the 5′ region. Nucleic Acids Res 16:6147–6156
Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, Hutchison III CA (1986) The sequence of a large L1Md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol 6:168–182
Martin SL, Voliva CF, Hardies SC, Edgell MH, Hutchison III CA (1985) Tempo and mode of concerted evolution in the L1 repeat family of mice. Mol Biol Evol 2:127–140
Matera GA, Hellmann U, Hintz MF, Schmid CW (1990) Recently transposed Alu repeats result from multiple source genes. Nucleic Acids Res 18:6019–6023
Mathias SL, Scott AF, Kazazian Jr HH, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254:1808–1810
Nei M, Gojobori T (1986) Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3:418–426
Price DK, Ayres JA, Pasqualone D, Cabell CH, Miller W, Hardison RC (1992) The 5′ ends of LINE1 repeats in rabbit DNA define subfamilies and reveal a short sequence conserved between rabbits and humans. Genomics 14:320–331
Quentin Y (1988) The Alu family developed through successive waves of fixation closely connected with primate lineage history. J Mol Evol 27:194–202
Saiki RK, Gelfand DH, Stoeffel S, Scharf SJ, Jiguchi R, Horn GT, Mullis KB, Erlich HA (1988) Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sanger F, Nichlen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 74:5463–5468
Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O'Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L (1987) Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113–125
Severynse DM, Hutchison III CA, Edgell MH (1992) Identification of transcriptional regulatory activity within the 5′ A-type monomer sequence of the mouse LINE-1 retroposon. Mammalian Genome 2:41–50
Shen MR, Batzer MA, Deininger PL (1991) Evolution of the master Alu gene(s). J Mol Evol 33:311–320
Skowronski J, Singer MF (1986) The abundant LINE-1 family of repeated DNA sequences in mammals: genes and pseudogenes. Cold Spring Harbor Symp Quant Biol 51:457–463
Slagel V, Flemington E, Traina-Dorge V, Bradshaw H, Deininger P (1987) Clustering and subfamily relationships of the Alu family in the human genome. Mol Biol Evol 4:19–29
Sneath PHA, Sokal RR (1973) Numerical taxonomy: the principles and practice of numerical classification. Freeman, San Francisco, CA
Stanhope MJ, Czelusniak J, Si J-S, Nickerson J, Goodman M (1992) A molecular perspective on mammalian evolution from the gene encoding interphotoreceptor retinoid binding protein, with convincing evidence for bat monophyly. Mol Phyl Evol 1:148–160
Stanhope MJ, Bailey WJ, Czelusniak J, Goodman M, Si J-S, Nickerson J, Sgouros JG, Singer GAM, Kleinschmidt TK (1993) A molecular view of primate supraordinal relationships from the analysis of both nucleotide and amino acid sequences. In: MacPhee R (ed) Primates and their relatives in phylogenetic perspective. Plenum, New York pp 251–292
Tagle DA, Stanhope MJ, Siemieniak DR, Benson P, Goodman M, Slightom JL (1992) The β-globin gene cluster of the prosimian primateGalago crassicaudatus: nucleotide sequence determination of the 41-kb cluster and comparative sequence analyses. Genomics 13:741–760
Willard C, Nguyen HT, Schmid CW (1987) Existence of at least three distinct Alu subfamilies. J Mol Evol 26:180–186
Xiong Y, Eickbush TH (1988) The site-specific ribosomal DNA insertion element R1Bm belongs to a class of non-long-terminal-repeat retrotransposons. Mol Cell Biol 8:114–123
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Stanhope, M.J., Tagle, D.A., Shivji, M.S. et al. Multiple L1 progenitors in prosimian primates: Phylogenetic evidence from ORF1 sequences. J Mol Evol 37, 179–189 (1993). https://doi.org/10.1007/BF02407354
Issue Date:
DOI: https://doi.org/10.1007/BF02407354