Summary
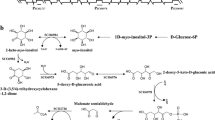
Fructose-1,6-bisphosphatase is a key enzyme in gluconeogenesis and the FBP1 gene is not transcribed during growth with glucose. Genetic analysis indicated a positive regulation of FBP1 expression after exhaustion of glucose. By linker-deletion analysis, two upstream activation sites (UAS1 and UAS2) were localized and the respective UAS-binding factors (DAP I and DAP II for derepression activating protein) were identified by gel retardation. UAS1 and UAS2 span about 30 bp each, and are separated by approximately 30 bp. Both UAS sites act synergistically. Although UAS1 showed some similarities to the DNA-binding consensus for the general yeast activator Rap1, competition experiments and DEAE-chromatography proved that DAP I and Rap1 correspond to different proteins. Gel retardation by DAP I depended on carbon sources and did not occur in cells growing logarithmically with glucose, whereas a strong retardation signal was obtained with ethanol-grown cells. The present results suggest that DAP I and DAP II are the final regulatory elements for glucose derepression.
Similar content being viewed by others
References
Arcangiolo B, Lescure B (1985) EMBO J 4:2627–2633
Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (eds.) (1987–1989) Current protocols in molecular biology. John Wiley and Sons, New York
Bailey RB, Woodward A (1984) Mol Gen Genet 193:507–512
Barnett JA, Kornberg HL (1960) J Gen Microbiol 23:65–82
Bartrons R, van Schaftingen E, Vissers S, Hers HG (1982) FEBS Lett 143:137–140
Bradford M (1976) Anal Biochem 72:248–254
Buchmann AR, Kimmerly WJ, Rine J, Kornberg RD (1988a) Mol Cell Biol 8:210–225
Buchmann AR, Lue NF, Kornberg RD (1988b) Mol Cell Biol 8:5086–5099
Burlini N, Morandi S, Pellegrini R, Tortora P, Guerritore A (1987) Biophys Acta 1014:153–161
Carlson M (1987) J Bacteriol 169:4873–4877
Carlson M, Osmond BC, Botstein D (1981) Genetics 98:25–40
Carlson M, Osmond BC, Neigeborn L, Botstein D (1984) Genetics 107:19–32
Celenza JL, Carlson M (1986) Science 233:1175–1180
Celenza JL, Eng FJ, Carlson M (1989) Mol Cell Biol 9:5045–5054
Ciriacy M (1975) Mutat Res 29:315–326
Ciriacy M (1977) Mol Gen Genet 154:213–220
Entian K-D (1980) Mol Gen Genet 178:633–637
Entian K-D (1986) Microbiol Sci 3:366–371
Entian K-D, Mecke D (1982) J Biol Chem 257:870–874
Entian K-D, Zimmermann FK (1980) Mol Gen Genet 177:345–350
Entian K-D, Zimmermann FK (1982) J Bacteriol 151:1123–1128
Entian K-D, Zimmermann FK, Scheel I (1977) Mol Gen Genet 156:99–105
Entian K-D, Vogel RF, Rose M, Hofmann L, Mecke D (1988) FEBS Lett 236:195–200
Ephrussi B, Slominski PP, Yoshio Y, Tavlitzki J (1956) Compt Rend Lab Carlsberg Sér Physiol 26:87–102
Fernandez E, Moreno F, Rodicio R (1992) Eur J Biochem 204:983–990
Fields S, Song O (1989) Nature 340:245–246
Fujita A, Matsumoto S, Kuhara S, Misumi Y, Kobayashi H (1990) Gene 89:93–99
Gancedo C (1971) J Bacteriol 107:401–405
Gancedo JM (1992) Eur J Biochem (in press)
Gancedo JM, Gancedo C (1986) FEMS Microbiol Rev 32:179–187
Gancedo C, Schwerzmann K (1976) Arch Microbiol 109:221–225
Gancedo C, Giner A, Salas ML, Sols A (1965) Biochem Biophys Res Commun 20:15–20
Gancedo C, Gancedo JM, Sols A (1967) Biochem Biophys Res Commun 26:528–531
Gascón S, Neumann NP, Lampen JO (1968) J Biol Chem 243:1573–1577
Guarente L (1983) Methods Enzymol 101:181–191
Haarasilta S, Oura E (1975) Eur J Biochem 52:1–7
Harashima S, Miller AM, Tanada K, Kusumoto K, Tanaka K, Mukai Y, Nasmyth K, Oshima Y (1989) Mol Cell Biol 9:4523–4530
Ito H, Fukuda Y, Marata K, Kimura A (1983) J Bacteriol 153:163–168
Lederer B, Vissers S, van Schaftingen E, Hers H-G (1981) Biochem Biophys Res Commun 103:1281–1287
Lenz AG, Holzer H (1980) FEBS Lett 109:271–274
Matsumoto K, Yoshimatsu T, Oshima Y (1983) J Bacteriol 153:1405–1414
Mazòn MJ, Gancedo JM, Gancedo C (1982) J Biol Chem 257:1128–1130
Melcher K, Entian K-D (1992) Curr Genet 21:295–300
Mercado JJ, Vincent O, Gancedo JM (1991) FEBS Lett 291:97–100
Michels C, Hahnenberger AKM, Sylvestre Y (1983) J Bacteriol 153:574–578
Müller D, Holzer H (1981) Biochem Biophys Res Commun 103:926–933
Myers AM, Tzagaloff A, Kinney DM, Lusty CJ (1986) Gene 45:299–310
Nehlin JO, Ronne H (1990) EMBO J 9:2891–2898
Nehlin JO, Carlberg M, Ronne H (1991) EMBO J 10:3373–3378
Neigeborn L, Carlson M (1984) Genetics 108:845–858
Neigeborn L, Carlson M (1987) Genetics 115:247–253
Niederacher D, Entian K-D (1987) Mol Gen Genet 206:505–509
Polakis ES, Bartley W (1965) Biochem J 97:284–297
Rose M, Entian K-D, Hofmann L, Vogel RF, Mecke D (1988) FEBS Lett 241:55–59
Rothstein RJ, Sherman F (1980) Genetics 94:871–889
Sambrock J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York
Schamhart DHJ, ten Berge AMA, van de Poll KW (1975) J Bacteriol 121:747–752
Schüller H-J, Entian K-D (1988) Gene 67:247–257
Schüller H-J, Entian K-D (1991) J Bacteriol 173:2045–2052
Sedivy JM, Fraenkel DG (1985) J Mol Biol 186:307–319
Shuster J, Yu J, Cox D, Chan RVL, Smith M, Young ET (1986) Mol Cell Biol 6:1894–1902
Spiegelman S, Dunn R (1947) J Gen Physiol 31:153–173
Strittmatter CF (1957) J Gen Microbiol 16:169–183
Swanson WH, Clifton CB (1948) J Bacteriol 56:115–124
Tautz D, Renz M (1983) Anal Biochem 132:14–19
Thrash-Bingham C, Fangman WL (1989) Mol Cell Biol 9:809–816
Wickner RB (1974) J Bacteriol 117:252–260
Wijk R van, Ouwehand J, van den Bos T, Koningsberger VV (1969) Biochim Biophys Acta 186:178–191
Witt I, Kronau R, Holzer H (1966) Biochim Biophys Acta 118:522–537
Zamenhoff S (1957) Methods Enzymol 3:696–704
Zhang M, Rosenblum-Vos LS, Lowry C, Boake KA, Zitomer RS (1991) Gene 97:153–161
Zimmermann FK, Eaton N (1974) Mol Gen Genet 134:261–272
Zimmermann FK, Scheel I (1977) Mol Gen Genet 154:75–82
Zimmermann FK, Kaufmann I, Rasenberger H, Haußmann P (1977) Mol Gen Genet 151:95–103
Author information
Authors and Affiliations
Additional information
Communicated by F. K. Zimmermann
Rights and permissions
About this article
Cite this article
Niederacher, D., Schüller, H.J., Grzesitza, D. et al. Identification of UAS elements and binding proteins necessary for derepression of Saccharomyces cerevisiae fructose-1,6-bisphosphatase. Curr Genet 22, 363–370 (1992). https://doi.org/10.1007/BF00352437
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00352437