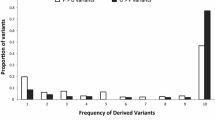
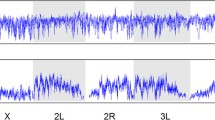
Abstract
Rates of synonymous and nonsynonymous substitution were investigated for 24 genes in three Drosophila species, D. pseudoobscura, D. subobscura, and D. melanogaster. D. pseudoobscura and D. subobscura, two distantly related members of the obscura clade, differ on average by 0.29 synonymous nucleotide substitutions per site. D. melanogaster differs from the two obscura species by an average of 0.81 synonymous substitutions per site. Using a method developed by Gillespie, we investigated the variance to mean ratio, or Index of Dispersion, R, of substitutions along the three species' branches to test the fundamental prediction of the neutral theory of molecular evolution, E(R) = 1. For nonsynonymous substitutions, the average R, Ra is 1.6, which is not significantly different from the neutral theory prediction. Only 5 of the 24 genes had significantly large Ra valves, and 12 of the genes had Ra estimates of less than one. In contrast, the Index of Dispersion for synonymous substitutions was significantly large for 12 of the 24 genes, with an average of Rs = 4.4, also statistically significant. These findings contrast with results for mammals, which showed overdispersion of nonsynonymous substitutions, but not of synonymous substitutions. Weak selection acting to maintain codon bias in Drosophila, but not in mammals, may be important in explaining the high variance in the rate of synonymous substitutions in this group of organisms.
Similar content being viewed by others
References
Akashi, H., 1994. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics 136: 927-935.
Akashi, H., 1995. Inferring weak selection from patterns of polymorphism and divergence at ‘silent’ sites in Drosophila DNA. Genetics 139: 1067-1076.
Benson, A.R., 1995. The molecular evolution of the obscura group Chorion s15: A prominent role for codon bias. PhD thesis, Harvard University.
Britten, R.J., 1986. Rates of DNA sequence evolution differ between taxonomic groups. Science 231: 1393-1398.
Bulmer, M., 1989. Estimating the variability of substitution rates. Genetics 123: 615-619.
Bulmer, M., K.H. Wolfe, & P.M. Sharp, 1991. Synonymous nucleotide substitution rates in mammalian genes: implications for the molecular clock and the relationship ofmammalian orders. Proc. Natl. Acad. Sci. USA 88: 5974-5978.
Chao, L. & D.E. Carr, 1993. The molecular clock and the relationship between population size and generation time. Evolution 47: 688-690.
Comeron, J.M., 1995. A method for estimating the numbers of synonymous and nonsynonymous substitutions per site. J. Mol. Evol. 41: 1152-1159.
Comeron, J.M., 1997. Estudi de la variabilitat nucleotidica a Drosophila: RegiÛ Xdh a D. subobscura. PhD thesis. Barcelona, Spain. Universitat de Barcelona.
Easteal, S., 1988. Rate constancy of globin gene evolution in placental mammals. Proc. Natl. Acad. Sci. 85: 7622-7626.
Easteal, S., 1990. The pattern of mammalian evolution and the relative rate test of molecular evolution. Genetics 124: 165-173.
Easteal, S. & C. Collet, 1994. Consistent variation in amino-acid substitution rate, despite uniformity of mutation rate: Protein evolution in mammals is not neutral. Mol. Biol. Evol. 11: 643- 647.
Gillespie, J.H., 1984. The molecular clock may be an episodic clock. Proc. Natl. Acad. Sci. USA 81: 8009-8013.
Gillespie, J.H., 1986a. Variability of evolutionary rates of DNA. Genetics 113: 1077-1091.
Gillespie, J.H., 1986b. Rates of molecular evolution. Annu. Rev. Ecol. Syst. 17: 637-665.
Gillespie, J.H., 1987. Molecular evolution and the neutral allele theory. Oxford Surveys Evol. Biol. 4: 10-37.
Gillespie, J.H., 1989. Lineage effects and the index of dispersion of molecular evolution. Mol. Biol. Evol. 6: 636-647.
Gillespie, J.H., 1991. The Causes of Molecular Evolution. Oxford series in Ecology and evolution. Oxford University Press. New York.
Gillespie, J.H., 1993. Substitution processes in molecular evolution. I. Uniform and clustered substitutions in haploid model. Genetics 134: 971-981.
Gillespie, J.H., 1994a. Substitution processes in molecular evolution. II. Exchangeable models from population genetics. Evolution 48: 1101-1113.
Gillespie, J.H., 1994b. Substitution processes in molecular evolution. III. Deleterious alleles. Genetics 138: 943-952.
Goddard, K., A. Caccone & J.R. Powell, 1990. Evolutionary implications of DNA divergence in the Drosophila obscura group. Evolution 44: 1656-1670.
Jukes, T. H., & C. R. Cantor, 1969. Evolution of protein molecules. pp. 21-132 in Mammalian Protein Metabolism III, edited by H. N. Munro. Academic Press, New York.
Kimura, M., 1969. The rate of molecular evolution considered from the standpoint of population genetics. Proc. Natl. Acad. Sci. USA 63: 1181-1188.
Kimura, M., 1983. The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambridge.
Kimura, M. & T. Ohta, 1971. On the rate of molecular evolution. J. Molec. Evol. 1: 1-17
King, J.L. & T.H. Jukes, 1969. Non-Darwinian evolution. Science 164: 788-798.
Kreitman, M., 1983. Nucleotide polymorphism at the alcohol dehydrogenase locus of Drosophila melanogaster. Nature 304: 412-417.
Langley, C.H. & W.M. Fitch, 1973. The constancy of evolution: a statistical analysis of the α and β aemoglobins, cytochrome c, and fibrinipeptide A, pp. 246-262 in Genetic Structure of Populations, edited by N.E. Morton, Univ. of Hawaii Press, Honolulu.
Langley, C.H. & W.M. Fitch, 1974. An estimation of the constancy of the rate of molecular evolution. J. Mol. Evol. 3: 161-177.
Li, W.-H., 1997. Molecular evolution. Sinauer Assoc., Inc.
Li, W.-H. & D. Graur, 1991. Fundamentals of Molecular Evolution. Sinauer Assoc., Inc., Sunderland.
Li, W.-H., M. Gouy, P. M. Sharp, C. O'Huigin & Y.-W. Yang, 1990. Molecular phylogeny of Rodentia, Lagomorpha, Primates, Artiodactyla, and Carnivora and molecular clocks. Proc. Natl. Acad. Sci. USA 87: 6703-6707.
Li, W.-H., M. Tanimura & P. M. Sharp, 1987. An evaluation of the molecular clock hypothesis using mammalian DNA sequences. J. Mol. Evol. 25: 330-342.
Margoliash, E., 1963. Primary structure and evolution of cytochrome c. Proc. Natl. Acad. Sci. USA 50: 672-679.
Martin, A. P., & S. R. Palumbi, 1993. Body size, metabolic rate, generation time, and the molecular clock. Proc. Natl. Acad. Sci. USA 90: 4087-4091.
Nei, M. & D. Graur, 1984. Extent of protein polymorphism and the neutral mutation theory. Evol. Biol. 17: 73-118.
Ohta, T., 1973. Slightly deleterious mutant substitutions in evolution. Nature 246: 96-98.
Ohta, T., 1991. Multigene families and the evolution of complexity. J. Mol. Evol. 33: 34-41.
Ohta, T., 1992. The nearly neutral theory of molecular evolution. Annu. Rev. Ecol. Syst. 23: 263-286.
Ohta, T., 1993. An examination of the generation-time effect on molecular evolution. Proc. Natl. Acad. Sci. USA 90: 10676-10680.
Ohta, T., 1995. Synonymous and nonsynonymous substitutions in mammalian genes and the nearly neutral theory. J. Mol. Evol. 40: 56-63.
Ohta, T. & M. Kimura, 1971. On the constancy of the evolutionary rate of cistrons. J. Mol. Evol. 1: 18-25.
Riley, M., M.E. Hallas & R.C. Lewontin, 1989. Distinguishing the forces controlling variation at the Xdh locus in Drosophila pseudoobscura. Genetics 123: 359-369.
Sarich, V.M. & A.C. Wilson, 1973. Generation time and genomic evolution in Primates. Science 179: 1144-1147.
Schaeffer, S.W., C.F. Aquadro & W.W. Anderson, 1987. Restriction-map variation in the alcohol dehydrogenase region of Drosophila pseudoobscura. Mol. Biol. Evol. 4: 254-265.
Sharp, P.M., & W.-H. Li, 1989. On the rate of DNA sequence evolution in Drosophila. J. Mol. Evol. 28: 398-402.
Takahata, N. & M. Kimura, 1979. Genetic variability maintained in a finite population under mutation and autocorrelated random fluctuation of selection intensity. Proc. Natl. Acad. Sci. USA 76: 5813-5817.
Takahata, N., K. Iishi & H. Matsuda, 1975. Effect of temporal fluctuation of selection coefficient on gene frequency in a population. Proc. Natl. Acad. Sci. USA 72: 4541-4545.
Thomson, J.D., D.G. Higgins & T.J. Gibson, 1994. CLUSTAL W: improving the sensitivity of progressive sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680.
Wang, D., J.L. Marsh & F.J. Ayala, 1996. Evolutionary changes inthe expression pattern of a developmentally essential gene in three Drosophila species. Proc. Natl. Acad. Sci. USA 93: 7103-7107.
Wallis, M., 1996. The molecular evolution of vertebrate growth hormones: A pattern of near-stasisinterrupted by sustained bursts of rapid change. J. Mol. Evol. 43: 93-100.
Wells, R.S., 1996. Nucleotide variation at the Gpdh locus in the genus Drosophila. Genetics 143: 375-384.
Wright, F., 1990. ‘The effective number of codons’ used in a gene. Gene 87: 23-39.
Wu, C.-I. & W.-H. Li, 1985. Evidence for higher rates of nucleotide substitution in rodents than in man. Proc. Natl. Acad. Sci. USA 82: 1741-1745.
Zeng, L.-W. & M. Kreitman, 1996a. Simple strategy for sequencing cDNA clones. Biotechniques 1996 Sep; 21(3): 446-452
Zeng, L.-W. & M. Kreitman, 1996b. Rapid and cost-effective DNA sequencing strategy for PCR products. Trends in Genetics, Technical Tips Online #TL10017.
Zuckerkandl, E. & L. Pauling, 1962. Molecular disease, evolution, and genetic heterogeneity, pp. 189-225 in Horizons in Biochemistry, edited by M. Kasha and B. Pullman. Academic Press. New York.
Zuckerkandl, E. & L. Pauling, 1965. Evolutionary divergence and convergence in proteins, pp. 97-166 in Evolving Genes and Proteins, edited by V. Bryson and H.J. Vogel. Academic Press. New York.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zeng, LW., Comeron, J.M., Chen, B. et al. The molecular clock revisited: the rate of synonymous vs. replacement change in Drosophila. Genetica 102, 369–382 (1998). https://doi.org/10.1023/A:1017035109224
Issue Date:
DOI: https://doi.org/10.1023/A:1017035109224