Abstract
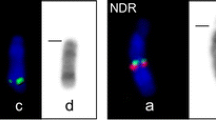
A combination of chromosomal banding and fluorescence in situ hybridization (FISH) was used to characterize the karyotype of Boselaphus tragocamelus (nilgai) relative to the domestic cattle standard karyotype. G-, Q- and C-band karyotypes of nilgai are presented, and the chromosomal complement of nilgai is determined to be 2n = 46 (female FN = 60, male FN = 59; NAA = 56), consistent with previous reports for the species. Comparisons with cattle identified extensive monobrachial homologies with some noteworthy exceptions. Chromosome 25 is centrically fused to 24, and chromosome 16 is acrocentric. Both appear to have additional pericentromeric material not seen in the equivalent cattle acrocentrics. This pericentromeric chromatin may be the result of de novo additions or translocation of pericentromeric material from chromosome 6, which is shown to be centrically fused to 13 but is only about two-thirds the length of cattle 6. Comparisons with cattle demonstrated that nilgai chromosome 17 has undergone a paracentric inversion and that chromosome 20 has two blocks of interstitial constitutive heterochromatin. The identities of both chromosomes were confirmed by chromosomal FISH. Furthermore, chromosomal banding and FISH were used to determine that autosome 14 has been fused to the ancestral X and Y of nilgai to form compound neo-X and -Y chromosomes. Additional FISH analyses were conducted to confirm other proposed chromosome homologies and to identify nucleolar organizing regions within the nilgai complement.
Similar content being viewed by others
References
Baker RJ, Davis SK, Bradley RD, Hamilton MJ, Van Den Bussche RA (1989) Ribosomal-DNA, mitochondrial-DNA, chromosomal, and allozymic studies on a contact zone in the pocket gopher, Geomys. Evolution 43: 63–75.
Barendse W, Vaiman D, Kemp SJ et al. (1997) A medium-density genetic linkage map of the bovine genome. Mammal Genome 8: 21–28.
Buckland RA, Evans HJ (1978a) Cytogenetic aspects of phylogeny in the Bovidae I. G-banding. Cytogenet Cell Genet 21: 42–63.
Buckland RA, Evans HJ (1978b) Cytogenetic aspects of phylogeny in the Bovidae II. C-banding. Cytogenet Cell Genet 21: 64–71.
Cai L, Taylor JF, Wing RA, Gallagher DS, Woo S-S, Davis SK (1995) Construction and characterization of a bovine bacterial artificial chromosome library. Genomics 29: 413–425.
Davis SK (1986) Population structure and patterns of speciation in Geomys (Rodentia: Geomyidae): An analysis using mitochondrial and ribosomal DNA. PhD Dissertation Washington University, Saint Louis, MO, USA.
Effron M, Bogart MH, Kumamoto AT, Benirschke K (1976) Chromosome studies in the mammalian subfamily Antilopinae. Genetica 46: 419–444.
Evans HJ, Buckland RA, Sumner AT (1973) Chromosome homology of heterochromatin in goat, sheep, and ox studied by banding techniques. Chromosoma 42: 383–402.
Gallagher DS Jr, Womack JE (1992) Chromosome conservation in the Bovidae. J Hered 83: 287–298.
Gallagher DS Jr, Ryan AM, Liou LS, Sastry KN, Womack JE (1993) Somatic cell mapping of conglutinin (CGN1) to cattle syntenic group U29 and fluorescence in situ localization to chromosome 28. Mammal Genome 4: 716–719.
Gallagher DS Jr, Derr JN, Womack JE (1994) Chromosome conservation among the advanced pecorans and determination of the primitive bovid karyotype. J Hered 85: 204–210.
Gallagher DS Jr, Ryan AM, Diamond G, Bevins CL, Womack JE (1995) Somatic cell mapping of β-defensin genes to cattle syntenic group U25 and fluorescence in situ localization to chromosome 27. Mammal Genome 6: 554–556.
Hansen KM (1973) Heterochromatin (C-bands) in bovine chromosomes. Hereditas 73: 65–70.
Iannuzzi L, Di Berardino D, Gustavsson I, Ferrara L, Di Meo GP (1987) Centromeric loss in translocations of centric fusion type in cattle and river buffalo. Hereditas 106: 73–81.
Iannuzzi L, Gallagher DS, Di Meo GP, Diamond G, Bevins CL, Womack JE (1996). High resolution FISH mapping of β-defensin genes in river buffalo and sheep chromosomes suggests a chromosome discrepancy in cattle standard karyotypes. Cytogenet Cell Genet 75: 10–13.
ISCNDA (1990) International system for cytogenetic nomenclature of domestic animals, Di Berardino D, Hayes H, Fries R, Long S (eds). Cytogenet Cell Genet 53: 65–79.
IUCN (1996) Red List of Threatened Animals, Baillie J & Groombridge B (eds). Gland, Switzerland: IUCN.
Kumamoto AT, Charter SC, Houck ML, Frahm M (1996) Chromosomes of Damaliscus (Artiodatyla, Bovidae): simple and complex centric fusion rearrangements. Chrom Res 4: 614–621.
MacDonald DW (1987) The Encyclopedia of Mammals. New York: Facts on File.
Modi WS, Gallagher DS, Womack JE (1996) Evolutionary histories of highly repeated DNA families among the Artiodactyla (Mammalia). J Mol Evol 42: 337–349.
Nowak RM (1991) Walker's Mammals of the World, 5th edn, Vol. II. Baltimore: Johns Hopkins University Press.
Petit P, Vermeesch JR, Marynen P, De Meurichy W (1994) Comparative cytogenetic study in the subfamily Tragelaphinae. Proceedings 11th European Colloquism on the Cytogenetics of Domestic Animals, pp. 109–113.
Pinkel D, Staume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitive, fluorescence hybridization. Proc Natl Acad Sci USA 83: 2934–2938.
Popescu CP, Long S, Riggs P, Womack JE, Schmutz S, Fries R, Gallagher DS (1996) Standardization of cattle chromosome nomenclature: Report of the committee for the standardization of the cattle karyotype. Cytogenet Cell Genet 74: 259–261.
Qumsiyeh MB, Baker RJ (1988) Comparative cytogenetics and the determination of primitive karyotypes. Cytogenet Cell Genet 47: 100–103.
Skow LC, Snaples SN, Davis SK, Taylor JF, Huang B, Gallagher DH (1996) Localization of bovine lymphocyte antigen (BoLA) DYA and class I loci to different regions of chromosome 23. Mammal Genome 7: 388–389.
Wilson DE, Reeder DM (1993) Mammal Species of the World, A Taxonomic and Geographic Reference, 2nd edn. Washington DC: Smithsonian Institution Press.
Wurster DH, Benirschke K (1968) Chromosome studies in the superfamily Bovoidea. Chromosoma 25: 152–171.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Gallagher, D.S., Davis, S.K., De Donato, M. et al. A Karyotypic Analysis of Nilgai, Boselaphus Tragocamelus (Artiodactyla: Bovidae). Chromosome Res 6, 505–514 (1998). https://doi.org/10.1023/A:1009268917856
Issue Date:
DOI: https://doi.org/10.1023/A:1009268917856