Abstract
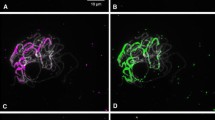
Tandem repetitive sequences consisting of 140-bp repetitive units were cloned from sugar cane genomic DNA and designated the SCEN family. In situ hybridization revealed that they were located on the centromeric region of almost all of the chromosomes of sugar cane. The 140-bp sequence included three CENP-B box-like sequences. Phylogenetic analysis of the members of the SCEN family revealed that the sequences had 75% homology with each other, on average, and that the sequences could not be further classified into smaller subfamilies. The copy number of the sequence was estimated to be 2.6 × 105 per haploid sugar cane genome and, therefore, 4.6 × 103 or 630 kb per chromosome on average.
Similar content being viewed by others
References
Aragón-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G (1996) A cereal centromeric sequence. Chromosoma 105: 261–268.
Bennett MD, Smith JB (1976) Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci 274: 227–274.
Brandes A, Thompson H, Dean C, Heslop-Harrison J (1997) Multiple repetitive DNA sequences in the paracentromeric regions of Arabidopsis thaliana L. Chrom Res 5: 238–246.
Choo KHA (1997) The Centromere. Oxford: Oxford University Press.
D'Hont A, Rao PS, Feldmann P, Grivet L, Islam-Faridi N, Taylor P, Glaszmann C (1995) Identification and characterization of sugar cane intergeneric hybrids, Saccharum officinarum x Erianthus arundinaceus, with molecular markers and DNA in situ hybridization. Theor Appl Genet 91: 320–326.
D'Hont A, Grivet L, Feldmann P, Rao S, Berding N, Glaszmann JC (1996) Characterization of the double genome structure of modern sugar cane cultivers (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet 250: 405–413.
Harrington JJ, Bokkelen GV, Mays RW, Gustashaw K, Willard HF (1997) Formation of de novo centromeres and construction of first-generation human artificial microchromosomes. Nature Genet 15: 345–355.
Heery DM, Gannon F, Powell R (1990) A simple method for subcloning DNA fragments from gel slices. Trends Genet 6: 173.
Jiang J, Nasuda S, Dong F, Scherrer CW, Woo S, Wing RA, Gill BS, Ward DC (1996) A conserved repetitive DNA element located in the centromeres of cereal chromosomes. Proc Natl Acad Sci USA 93: 14210–14213.
Kamm A, Schmidt T, Heslop-Harrison JS (1994) Molecular and physical organization of highly repetitive, undermethylated DNA from Pennisetum glaucum. Mol Gen Genet 244: 420–425.
Kipling D (1995) The Telomere. Oxford: Oxford University Press.
Murata M, Ogura Y, Motoyoshi F (1994) Centromeric repetitive sequences in Arabidopsis thaliana. Jpn J Genet 69: 361–370.
Murray HG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8: 4321–4325.
Moore G, Roberts M, Aragon-Alcaide L, Foote T (1997) Centromeric sites and cereal chromosome evolution. Chromosoma 105: 321–323.
Nagaki K, Tsujimoto H, Isono K, Sasakuma T (1995) Molecular characterization of a tandem repeat, Afa family, and distribution among Triticeae. Genome 38: 479–486.
Orgel LE, Crick FHC (1980) Selfish DNA: the ultimate parasite. Nature 284: 604–607.
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
Sart DD, Cancilla MR, Earle E, Mao J, Saffery R, Tainton KM, Kalitsis P, Martin J, Barry AE, Choo KHA (1997) A functional neo-centromere formed through activation of a latent human centromere and consisting of non-alpha-satellite DNA. Nature Genet 16: 144–153.
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425.
Thompson JD, Higgins1, DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680.
Willard HF (1990) Centromeres of mammalian chromosomes. Trends Genet 6: 410–415.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Nagaki, K., Tsujimoto, H. & Sasakuma, T. A Novel Repetitive Sequence of Sugar Cane, SCEN Family, Locating on Centromeric Regions. Chromosome Res 6, 295–302 (1998). https://doi.org/10.1023/A:1009270824142
Issue Date:
DOI: https://doi.org/10.1023/A:1009270824142