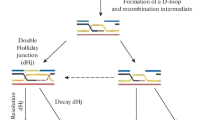
Abstract
Finding the right partner is a central problem in homologous recombination. Common to all models for general recombination is a homologous pairing and DNA strand exchange step. In prokaryotes this process has mainly been studied with the RecA protein ofEscherichia coli. Two approaches have been used to find homologous pairing and DNA strand exchange proteins in eukaryotes. A biochemical approach has resulted in numerous proteins from various organisms. Almost all of these proteins are biochemically fundamentally different from RecA. The in vivo role of these proteins is largely not understood. A molecular-genetical approach has identified structural homologs to theE. coli RecA protein in the yeastSaccharomyces cerevisiae and subsequently in other organisms including other fungi, mammals, birds, and plants. The biochemistry of the eukaryotic RecA homologs is largely unsolved. For the fungal RecA homologs (S. cerevisiae RAD51, RAD55, RAD57, DMC1; Schizosaccharomyces pombe rad51; Neurospora crassa mei3) a role in homologous recombination and recombinational repair is evident. Besides recombination, homologous pairing proteins might be involved in other cellular processes like chromosome pairing or gene inactivation.
Similar content being viewed by others
References
Aboussekhra, A., Chanet, R., Adjiri, A., and Fabre, F., Semidominant suppressors of Srs2 helicase mutations ofSaccharomyces cerevisiae map in theRAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Molec. cell. Biol.12 (1992) 3224–3234.
Arai, N., Kawasaki, K., and Shibata, T., A multi-component protein of a fission yeast which promotes joint molecule formation from homologous DNAs. J. biol. Chem.267 (1992) 3514–3522.
Bähler, J., Hagens, G., Holzinger, G., Scherthan, H., and Heyer, W.-D.,Saccharomyces cerevisiae cells lacking the homologous pairing protein p175SEP1 arrest at pachytene during meiotic prophase. Chromosoma. In press.
Basile, G., Aker, M., and Mortimer, R. K., Nucleotide sequence and transcriptional regulation of the yeast recombinational repair geneRAD51. Molec. cell. Biol.12 (1992) 3235–3246.
Bezzubova, O., Shinohara, A., Mueller, R. G., Ogawa, H., and Buerstedde, J.-M., A chickenRAD51 homologue is expressed at high levels in lymphoid and reproductive organs. Nucl. Acids Res.21 (1993) 1577–1580.
Bishop, D. K., Park, D., Xu, L., and Kleckner, N.,DMC1, A meiosis-specific yeast homolog ofE. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell69 (1992) 439–456.
Bollag, R. J., Elwood, D. R., Tobin, E. D., Godwin, A. R., and Liskay, R. M., Formation of heteroduplex DNA during mammalian intra-chromosomal gene conversion. Molec. cell. Biol.12 (1992) 1546–1552.
Bortner, C., Hernandez, T. R., Lehman, I. R., and Griffith, J., Herpes simplex virus 1 single-stranded DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J. molec. Biol.231 (1993) 241–250.
Cerutti, H., Osman, M., Grandoni, P., and Jagendorf, A. T., A homolog ofEscherichia coli RecA protein in plastids of higher plants. Proc. natl Acad. Sci. USA89 (1992) 8068–8072.
Cerutti, H., and Jagendorf, A. T., DNA strand transfer activity in Pea (Pisum sativum L.) chloroplasts. Plant Physiol.102 (1993) 145–153.
Cerutti, H., Ibrahim, H. Z., and Jagendorf, A. T., Treatment of Pea (Pisum sativum L.) protoplasts with DNA-damaging agents induces a 39-kilodalton chloroplast protein immunologically related toEscherichia coli RecA. Plant Physiol.102 (1993) 155–163.
Cheng, R., Baker, T. I., Cords, C. E., and Radloff, R. J.,mei-3, a recombination and repair gene ofNeurospora crassa, encodes a RecA-like protein. Mutation Res.294 (1993) 223–234.
Clark, A. B., Dykstra, C. C., and Sugino, A., Isolation, DNA sequence, and regulation of aSaccharomyces cerevisiae gene that encodes DNA strand transfer protein α. Molec. cell. Biol.11 (1991) 2576–2582.
Dykstra, C. C., Hamatake, R. K., and Sugino, A., DNA strand transfer protein β from yeast mitotic cells differs from strand transfer protein α from meiotic cells. J. biol. Chem.265 (1990) 10968–10973.
Dykstra, C. C., Kitada, K., Clark, A. B., Hamatake, R. K., and Sugino, A., Cloning and characterization of DST2, the gene for DNA strand transfer protein β fromSaccharomyces cerevisiae. Molec. cell. Biol.11 (1991) 2583–2592.
Egelman, E. H., What do X-ray crystallographic and electron microscopic structural studies of the RecA protein tell us about recombination? Curr. Opin. in struct. Biol.3 (1993) 189–197
Eggleston, A. K., and Kowalczykowski, S. C., An overview of homologous pairing and DNA strand exchange proteins. Biochimie73 (1991) 163–176.
Fields, S., and Song, O.-K., A novel genetic system to detect protein: protein interactions. Nature340 (1989) 245–246.
Fishel, R. A., Detmer, K., and Rich, A., Identification of homologous pairing and strand-exchange activity from a human tumor cell line based on Z-DNA affinity chromatography. Proc. natl Acad. Sci. USA85 (1988) 36–40.
Fisher, C., Parks, R. J., Lanzon, M. L., and Evans, D. H., Heteroduplex DNA formation is associated with replication and recombination in poxvirus infected cells. Genetics129 (1991) 7–18.
Fotheringham, S., and Holloman, W. K., Extrachromosomal recombination is deranged in therec2 mutant ofUstilago maydis. Genetics129 (1991) 1053–1060.
Friedberg, E. C., Deoxyribonucleic acid repair in the yeastSaccharomyces cerevisiae. Microbiol. Rev.52 (1988) 70–102.
Gao, M., and Knipe, D. M., Genetic evidence for multiple nuclear functions of the herpes simplex virus ICP8 DNA binding protein. J. Virol.63 (1989) 5258–5267.
Goyon, C., and Lichten, M., Timing of molecular events in meiosis inSaccharomyces cerevisiae: Stable heteroduplex DNA is formed in meiotic prophase. Molec. cell. Biol.13 (1993) 373–382.
Griffith, J. D., and Harris, L. D., DNA strand exchanges. CRC crit. Rev. Biochem.23 (1988) S43-S86.
Gu, L., Huang, S.-M., and Sander, M., Drosophila Rrp1 complementsE. coli xth nfo mutants: Protection against both oxidative and alkylation-induced DNA damage. Nucl. Acids Res.21 (1993) 4788–4795.
Halbrook, J., and McEntee, K., Purification and characterization of a DNA-pairing and strand transfer activity from mitoticSaacharomyces cerevisiae. J. biol. Chem.264 (1989) 21403–21412.
Hall, S. D., Kane, M. F., and Kolodner, R. D., Identification and characterization of theEscherichia coli recT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. J. Bact.175 (1993) 277–287.
Heyer, W.-D., and Kolodner, R. D., Enzymology of homologous recombination inSaccharomyces cerevisiae. Prog. nucl. Acid Res. molec. Biol.46 (1993) 221–271.
Hinnebusch, A. G., and Liebman, S. W., Protein synthesis and translational control inSaccharomyces cerevisiae, in: The Molecular and Cellular Biology of the Yeast Saccharomyces. Genome Dynamics, Protein Synthesis, and Energetics, pp. 627–735. Eds. J. R. Broach, J. Pringle and E. Jones, CSHL Press, Cold Spring Harbor 1992.
Holden, D. W., Spanos, A., and Banks, G. R., Nucleotide sequence of theREC1 gene ofUstilago maydis. Nucl. Acids Res.17 (1989) 10489.
Holliday, R., Altered recombination frequencies in radiation sensitive strains ofUstilago. Mutation Res.4 (1967) 275–288.
Howard-Flanders, P., West, S. C., and Stasiak, A., Role of RecA protein spiral filaments in genetic recombination. Nature309 (1984) 215–220.
Hsieh, P., Meyn, M. S., and Camerini-Otero, R. D., Partial purification and characterization of a recombinase from human cells. Cell44 (1986) 885–894.
Hsu, C. L., and Stevens, A., Yeast cells lacking 5′–3′ exoribonuclease I contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Molec. cell. Biol.13 (1993) 4826–4835.
Huang, K. N., and Symington, L. S., A 5′–3′ exonuclease fromSaccharomyces cerevisiae is required forin vitro recombination between linear DNA molecules with overlapping homology. Molec. cell. Biol.13 (1993) 3125–3134.
Johnson, A. W., and Kolodner, R. D., Strand exchange protein I fromSaccharomyces cerevisiae: a novel multifunctional protein that contains DNA strand exchange and exonuclease activites. J. biol. Chem.266 (1991) 14046–14054.
Johnson, A. W., and Kolodner, R. D., Characterization of the interaction ofSaccharomyces cerevisiae strand exchange protein I with DNA. J. biol. Chem. In press.
Johnson, A. W., and Kolodner, R. D., The activity of theSaccharomyces cerevisiae strand exchange protein 1 intrinsic exonuclease during joint molecule formation. J. biol. Chem. In press.
Kaus, J. A., and Mortimer, R. K., Nucleotide sequence of theRAD57 gene ofSaccharomyces cerevisiae. Gene105 (1991) 139–140.
Kawasaki, I., Sugano, S., and Ikeda, H., Calf thymus histone H1 is a recombinase that catalyzes ATP-independent DNA strand transfer. Proc. natl Acad. Sci. USA86 (1989) 5281–5285.
Kawasaki, I., Sugano, S., and Ikeda, H., Calf thymus histone H1 is a recombinase that catalyzes ATP-independent DNA strand transfer. Proc. natl Acad. Sci. USA87 (1989) 6128.
Kearsey, S., and Kipling, D., Recombination and RNA processing: a common strand? T Cell Biol.1 (1991) 110–112.
Kim, J., Genes controlling conjugation and mitotic cell division in yeastSaccharomyces cerevisiae. Ph. D. thesis (1988) Massachusetts Institute of Technology.
Kim, J., Ljungdahl, P. O., and Fink, G. R.,kem mutations affect nuclear fusion inSaccharomyces cerevisiae. Genetics126 (1990) 799–812.
Kipling, D., and Kearsey, S. E., TFIIS and strand-transfer proteins. Nature353 (1991) 509.
Kleckner, N., Padmore, R., and Bishop, D. K., Meiotic chromosome metabolism: one view. Cold Spring Harb. Symp. quant. Biol.56 (1991) 729–743.
Kmiec, E., and Holloman, W. K., Homologous pairing of DNA molecules promoted by a protein fromUstilago. Cell29 (1982) 367–374.
Kobayashi, T., Hotta, Y., and Tabata, S., Isolation and characterization of a yeast gene that is homologous with a meiosisspecific cDNA from a plant. Molec. gen. Genet.237 (1993) 225–232.
Kolodner, R., Evans, D. H., and Morrison, P. T., Purification and characterization of an activity fromSaccharomyces cerevisiae that catalyzes homologous pairing and strand exchange. Proc. natl Acad. Sci. USA84 (1987) 5560–5564.
Kowalczykowski, S. C., Biochemistry of genetic recombination: Energetics and mechanism of DNA strand exchange. A. Rev. Biophys. biophys. Chem.20 (1991) 539–575.
Lovett, S., and Mortimer, R. K., Characterization of null mutants of therad55 gene ofSaccharomyces cerevisiae: effects of temperature, osmotic strength, and mating type. Genetics116 (1987) 547–533.
Lovett, S. T., Sequence of the RAD55 gene ofSaccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene. In press.
Lowenhaupt, K., Sander, M., Hauser, C., and Rich, A., Drosophila melanogaster strand transferase. J. biol. Chem.264 (1989) 20568–20575.
McCarthy, J. G., Sander, M., Lowenhaupt, K., and Rich, A., Sensitive homologous recombination strand transfer assay: Partial purification of aDrosophila melanogaster enzyme and detection of sequence effects on the strand transfer activity of RecA protein. Proc. natl Acad. Sci. USA85 (1988) 5854–5858.
Meselson, M. S., and Radding, C. R., A general model for genetic recombination. Proc. natl Acad. Sci. USA72 (1975) 358–361.
Milne, G. T., and Weaver, D. T., Dominant negative alleles ofRAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev.7 (1993) 1755–1765.
Moore, S. P., and Fishel, R., Purification and characterization of a protein from human cells which promotes homologous pairing of DNA. J. biol. Chem.265 (1990) 11108–11117.
Moore, S. P., Erdile, L., Kelly, T., and Fishel, R., The human homologous pairing protein HPP-1 is specifically stimulated by the cognate single-stranded binding protein hRP-A. Proc. natl Acad. Sci. USA88 (1991) 9067–9071.
Morita, T., Yoshimura, Y., Yamamoto, A., Murata, K., Mori, M., Yamamoto, H., and Matsuhiro, A., A mouse homolog of theEscherichia coli recA andSaccharomyces cerevisiae Rad51 genes. Proc. natl Acad. Sci. USA90 (1993) 6577–6580.
Morrison, D. P., and Hastings, P. J., Characterization of the mutator mutationmut 5-1. Molec. gen. Genet.175 (1979) 57–65.
Muris, D. F. R., Vreken, K., Carr, A. M., Broughton, B. C., Lehmann, A. R., Lohman, P. H. M., and Pastink, A., Cloning ofRAD51 homolog ofSchizosaccharomyces pombe. Nucl. Acids Res.21 (1993) 4586–4591.
Nag, D. K., and Petes, T. D., Physical detection of heteroduplexes during meiotic recombination in the yeastSaccharomyces cerevisiae. Molec. cell. Biol.13 (1993) 2324–2331.
Nugent, M., Huang, S.-M., and Sander, M., Characterization of the apurinic endonuclease activity of Drosophila Rrpl. Biochemistry32 (1993) 11445–11452.
Ogawa, T., Shinohara, A., Ogawa, H., and Tomizawa, J., Functional structure of the RecA protein found by chimera analysis. J. molec. Biol.226 (1992) 651–660.
Ogawa, T., Yu, X., Shinohara, A., and Egelman, E. H., Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science259 (1993) 1896–1899.
Ogawa, T., Shinohara, A., Nabetani, A., Ikeya, T., Yu, X., Egelman, E. H., and Ogawa, H., RecA-like recombination proteins in Eukaryotes: functions ofRad51 andRad52 genes ofSaccharomyces cerevisiae. Cold Spring Harb. Symp. quant. Biol. In press.
Pang, Q., Hays, J. B., and Rajagopal, I., A plant cDNA that partially complementsEscherichia coli recA mutations predicts a polypeptide not strongly homologous to RecA proteins. Proc. natl Acad. Sci. USA89 (1992) 8073–8077.
Panyutin, I. G., and Hsieh, P., Formation of a single base mismatch impedes spontaneous DNA branch migration. J. molec. Biol.230 (1993) 413–424.
Petes, T. D., Malone, R. E., and Symington, L. S., Recombination in Yeast. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Genome Dynamics, Protein Synthesis, and Energetics. pp. 407–521. Eds J. R. Broach, J. Pringle, E. Jones. CSHL Press, Cold Spring Harbor 1992.
Radding, C. M., Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. biol. Chem.266 (1991) 5355–5358.
Roca, A. I., and Cox, M. M., The RecA protein: Structure and function. CRC crit. Rev.25 (1990) 415–456.
Saeki, T., Machida, I., and Nakai, S., Genetic control of diploid recovery after γ-irradiation in the yeastSaccharomyces cerevisiae. Mutation Res.73 (1980) 251–265.
Sander, M., Lowenhaupt, K., Lane, W. S., and Rich, A., Cloning and characterization of Rrpl, the gene encoding Drosophila strand transferase: carboxy-terminal homology to DNA repair endo/exonucleases. Nucl. Acids Res.19 (1991) 4523–4529.
Sander, M., Lowenhaupt, K., and Rich, A., Drosophila Rrpl protein: An apurinic endonuclease with homologous recombination activities. Proc. natl Acad. Sci. USA88 (1991) 6780–6784.
Shinohara, A., Ogawa, H., and Ogawa, T., Rad51 protein involved in repair and recombination inS. cerevisiae is a RecA-like protein. Cell69 (1992) 457–470.
Shinohara, A., Ogawa, H., Hatsuda, Y., Ushio, N., Ikeo, K., and Ogawa, T., Cloning of human, mouse and fission yeast recombination genes homologous to Rad51 and recA. Nature Genet.4 (1993) 239–243.
Shuster, E. O., and Byers, B., Pachytene arrest and other meiotic effects of the Start mutations inSaccharomyces cerevisiae. Genetics123 (1989) 29–43.
Sikorav, J.-L., and Church, G. M., Complementary recognition in condensed DNA: accelerated DNA renaturation. J. molec. Biol.222 (1991) 1085–1108.
Silberstein, Z., Shalit, M., and Cohen, A., Heteroduplex strand-specificity in restriction-stimulated recombination by the RecE pathway ofEscherichia coli. Genetics133 (1993) 439–448.
Smith, G. R., Homologous recombination in procaryotes. Microbiol. Rev.52 (1988) 1–28.
Stasiak, A., Three-stranded DNA structure: is this thesecret of DNA homologous recognition? Molec. Microbiol.6 (1992) 3267–3276.
Stevens, A., and Maupin, M. K., A 5′−3′ exoribonuclease ofSaccharomyces cerevisiae: size and novel substrate specificity. Archs Biochem. Biophys.252 (1987) 339–347.
Story, R. M., Weber, I. T., and Steitz, T. A., The structure of theE. coli recA protein monomer and polymer. Nature355 (1992) 318–325.
Story, R. M., and Steitz, T. A., Structure of the recA protein-ADP complex. Nature355 (1992) 374–376.
Story, R. M., Bishop, D. K., Kleckner, N., and Steitz T. A., Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science259 (1993) 1892–1896.
Sugino, A., Nitiss, J., and Resnick, M. A., ATP-independent DNA strand transfer catalyzed by protein(s) from meiotic cells of the yeastSaccharomyces cerevisiae. Proc. natl Acad. Sci. USA85 (1988) 3683–3687.
Svaren, J., Inagami, K., Lovegren, E., and Chalkley, R., DNA denatures upon drying after ethanol precipitation. Nucl. Acids Res.15 (1987) 8739–8754.
Szankasi, P., and Smith, G. R., A single-stranded DNA exonuclease fromSchizosaccharomyces pombe. Biochemistry31 (1992) 6769–6773.
Szostak, J. W., Orr-Weaver, T. L., Rothstein, R. J., and Stahl, F. W., The double-strand-break repair model for recombination. Cell33 (1983) 25–35.
Tartof, K. D., and Henikoff, S., Trans-sensing effects from Drosophila to humans. Cell65 (1991) 201–203.
Tishkoff, D., Johnson, A. W., and Kolodner, R. D., Molecular and genetic analysis of the gene encoding theSaccharomyces cerevisiae strand exchange protein SEP1. Molec. cell. Biol.11 (1991) 2593–2608.
Walker, J. E., Saraste, M., Runswick, M. J., and Gay, N. J., Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and the other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J.1 (1982) 945–951.
West, S. C., Enzymes and molecular mechanisms of genetic recombination. A. Rev. Biochem.61 (1992) 603–640.
Zhang, W., and Evans, D. H., DNA strand exchange catalyzed by proteins from vaccinia virus-infected cells. J. Virol.67 (1993) 204–212.
Yoshimura, Y., Morita, T., Yamamoto, A., and Matsuhiro, A., Cloning sequence of the human RecA-like gene cDNA. Nucl. Acids Res.21 (1993) 1665.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Heyer, W.D. The search for the right partner: Homologous pairing and DNA strand exchange proteins in eukaryotes. Experientia 50, 223–233 (1994). https://doi.org/10.1007/BF01924005
Published:
Issue Date:
DOI: https://doi.org/10.1007/BF01924005