Abstract
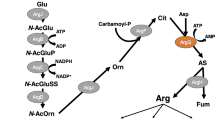
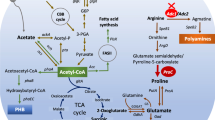
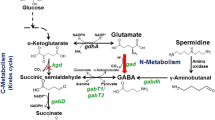
Arginine decarboxylase (ADC) is the first enzyme in the alternative route to putrescine in the polyamine biosynthesis pathway in bacteria and plants. In this study, we have focused on the effects of various types of short-term stresses on the transcript amount and specific activity of Synechocystis sp. PCC 6803 ADC. Our results reveal that the steady-state transcript accumulation and enzyme activity are not connected in a simple manner, since only photoheterotrophy and synergistic salt and high-light stress affected both parameters similarly. Changes in the steady-state ADC mRNA accumulation under the other short-term stress conditions studied had only a small impact on enzyme activity, suggesting post-translational regulation. Based on structural modeling, Synechocystis ADCs have a putative extra domain, which might be involved in the post-translational regulation of ADC activity in Synechocystis. In addition, two symmetric inter-subunit disulfide bonds seem to stabilize the dimeric structure of ADCs. There are two genes coding for ADC and agmatinase, another polyamine pathway enzyme, in Synechocystis genome, while the genes coding for ornithine decarboxylase and for some other enzymes in the polyamine pathway were not identified with homology searches.





Similar content being viewed by others
Abbreviations
- ADC:
-
Arginine decarboxylase
- AIH:
-
Agmatine iminohydrolase
- DapDC:
-
Diaminopimelate decarboxylase
- DCMU:
-
3-(3′,4′-Dichlorphenyl)-1,1-dimethylurea
- DFMO:
-
DL-α-difluoromethylornithine
- NCPAH:
-
N-carbamoylputrescine amidohydrolase
- ODC:
-
Ornithine decarboxylase
- PDB:
-
Protein data bank
- PLP:
-
Pyridoxal-5′-phosphate
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SAM:
-
S-adenosylmethionine
- SAMDC:
-
S-adenosylmethionine decarboxylase
References
Almrud JJ, Oliveira MA, Kern AD, Grishin NV, Phillips MA, Hackert ML (2000) Crystal structure of human ornithine decarboxylase at 2.1. A resolution: structural insights to antizyme binding. J Mol Biol 295:7–16
Bagni N (1989) Polyamines in plant growth and development. In: Bachrach U, Heimer YM (eds) The physiology of polyamines, 2nd edn. CRC Press, Boca Raton, pp 107–120
Bagni N, Tassoni A (2001) Biosynthesis, oxidation and conjugation of aliphatic polyamines in plants. Amino Acids 20:301–317
Balbo PB, Patel CN, Sell KG, Adcock RS, Neelakantan S, Crooks PA, Oliveira MA (2003) Spectrophotometric and steady-state kinetic analysis of the biosynthetic arginine decarboxylase of Yersinia pestis utilizing arginine analogues as inhibitors and alternative substrates. Biochemistry 42:15189–15196
Barton GJ (1993) ALSCRIPT a tool to format multiple sequence alignments. Protein Eng 1:37–40
Bell E, Malmberg RL (1990) Analysis of cDNA encoding arginine decarboxylase from oat reveals similarity to the Escherichia coli arginine decarboxylase and evidence of protein processing. Mol Gen Genet 224:431–436
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Bouchereau A, Aziz A, Larher F, Martin-Tanguy JM (1999) Polyamines and environmental challenges: recent development. Plant Sci 140:103–125
Bradford MM (1976) A rapid sensitive method for the quantization of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Chattopadhyay MK, Gupta S, Sengupta DN, Ghosh B (1997) Expression of arginine decarboxylase in seedlings of indica rice (Oryza sativa L.) cultivars as affected by salinity stress. Plant Mol Biol 34:477–483
Coleman CS, Hu G, Pegg AE (2004) Putrescine biosynthesis in mammalian tissues. Biochem J 379:849–855
Galloway GL, Malmberg RL, Price RA (1998) Phylogenetic utility of the nuclear gene arginine decarboxylase: an example from Brassicaceae. Mol Biol Evol 15:1312–1320
Gibon Y, Blaesing OE, Hannemann J, Carillo P, Höhne M, Hendriks JHM, Palacios N, Cross J, Selbig J, Stitt M (2004) A robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell 16:3304–3325
Gill RT, Katsoulakis E, Schmitt W, Taroncher-Oldenburg G, Misra J, Stephanopoulos G (2002) Genome-wide dynamic transcriptional profiling of the light-to-dark transition in Synechocystis sp. Strain PCC 6803. J Bacteriol 184:3671–3681
Gokulan K, Rupp B, Pavelka MS Jr, Jacobs WR Jr, Sacchettini JC (2003) Crystal structure of Mycobacterium tuberculosis diaminopimelate decarboxylase, an essential enzyme in bacterial lysine biosynthesis. J Biol Chem 278:18588–18596
Hanfrey C, Sommer S, Mayer MJ, Burtin D, Michael AJ (2001) Arabidopsis polyamine biosynthesis: absence of ornithine decarboxylase and the mechanism of arginine decarboxylase activity. Plant J 27:551–560
Hao YJ, Kitashiba H, Honda C, Nada K, Moriguchi T (2005) Expression of arginine decarboxylase and ornithine decarboxylase genes in apple cells and stressed shoots. J Exp Bot 56:1105–1115
He L, Nada K, Kasukabe Y, Tachibana S (2002) Enhanced susceptibility of photosynthesis to low-temperature photoinhibition due to interruption of chill-induced increase of S-adenosylmethionine decarboxylase activity in leaves of spinach (Spinacia oleracea L.). Plant Cell Physiol 43:196–206
Hofmann K, Stoffel W (1993) TMbase—a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler 374:166
Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to light. Plant Cell 13:793–806
Horyn O, Luhovvy B, Lazarow A, Daikhin Y, Nissim I, Yudkoff M, Nissim I (2005) Biosynthesis of agmatine in isolated mitochondria and perfused rat liver: studies with 15N-labeled arginine. Biochem J 388:419–425. DOI: 10.1042/BJ20041260
Illingworth C, Mayer MJ, Elliott K, Hanfrey C, Walton NJ, Michael AJ (2003) The diverse bacterial origins of the Arabidopsis polyamine biosynthetic pathway. FEBS Lett 549:26–30
Jackson LK, Brooks HB, Osterman AL, Goldsmith EJ, Phillips MA (2000) Altering the reaction specificity of eukaryotic ornithine decarboxylase. Biochemistry 39:11247–11257
Jansonius JN (1998) Structure, evolution and action of of vitamin B6-dependent enzymes. Curr Opin Struct Biol 8:759–769
Jantaro S, Mäenpää P, Mulo P, Incharoensakdi A (2003) Content and biosynthesis of polyamines in salt and osmotically stresses cells of Synechocystis sp. PCC 6803. FEMS Microbiol Lett 228:129–135
Johnson MS, Lehtonen JV (2000) Comparison of protein three-dimensional structures. In: Higgins D, Taylor W (eds) Bioinformatics: sequence, structure and databanks. Oxford University Press, Oxford UK, pp 15–50
Johnson MS, Overington JP (1993) A structural basis for the comparison of sequences: an evaluation of scoring methodologies. J Mol Biol 233:716–738
Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res 3:109–136
Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Co 290:339–348
Kasinathan V, Wingler A (2004) Effect of reduced arginine decarboxylase activity on salt tolerance and on polyamine formation during salt stress in Arabidopsis thaliana. Physiol Plant 121:101–107
Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24:946–950
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Lehtonen JV, Still DJ, Rantanen VV, Ekholm J, Björklund D, Iftikhar Z, Huhtala M, Repo S, Jussila A, Jaakkola J, Pentikäinen O, Nyrönen T, Salminen T, Gyllenberg M, Johnson MS (2004) BODIL: a molecular modeling environment for structure–function analysis and drug design. J Comp Aided Mol Des 18:401–419
Malmberg RL, Smith KE, Bell E, Cellino ML (1992) Arginine Decarboxylase of oats is clipped from a precursor into two polypeptides found in the soluble enzyme. Plant Physiol 100:146–152
Malmberg RL, Cellino ML (1994) Arginine decarboxylase of oats is activated by enzymatic cleavage into two polypeptides. J Biol Chem 269:2703–2706
Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol 277:505–524
Minocha R, Long S, Maki H, Minocha SC (1999) Assays for the activities of polyamine biosynthetic enzymes using intact tissues. Plant Physiol Biochem 37:597–603
Mohamed A, Jansson C (1989) Influence of light on accumulation of photosynthesis-specific transcripts in the cyanobacterium Synechocystis 6803. Plant Mol Biol 13:693–700
Mullineaux CW (2001) How do cyanobacteria sense and respond to light? Mol Microbiol 41:965–971
Muro-Pastor MI, Reyes JC, Florencio FJ (2001) Cyanobacteria perceive nitrogen status by sensing intracellular 2-oxoglutarate levels. J Biol Chem 276:38320–38328
Patel CN, Adcock RS, Sell KG, Oliveira MA (2004) Crystallization, X-ray diffraction and oligomeric characterization of arginine decarboxylase from Yersinia pestis, a key polyamine biosynthetic enzyme. Acta Cryst D 60:2396–2398
Primikirios NI, Roubelakis-Angelakis KA (1999) Cloning and expression of an arginine decarboxylase cDNA from Vitis viniferea L. cell-suspension cultures. Planta 208:574–582
Quintero MJ, Muro-Pastor AM, Herrero A, Flores E (2000) Arginine catabolism in the cyanobacterium Synechocystis sp. Strain PCC 6803 involves the urea cycle and arginase pathway. J Bacteriol 182:1008–1015
Ray SS, Bonanno JB, Rajashankar KR, Pinho MG, He G, De Lencastre H, Tomasz A, Burley SK (2002) Cocrystal structures of diaminopimelate decarboxylase: mechanism, evolution, and inhibition of an antibiotic resistance accessory factor. Structure 10:1499–1508
Rost B (1996) PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol 266:525–539
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17:355–362
Tassoni A, Fornalé S, Bagni N (2003) Putative ornithine decarboxylase activity in Arabidopsis thaliana: inhibition and intracellular localization. Plant Physiol Biochem 41:871–875
Urano K, Yoshiba Y, Nanjo T, Ito T, Yamaguchi-Shinozaki K, Shinozaki K (2004) Arabidopsis stress-inducible gene for arginine decarboxylase AtADC2 is required for accumulation of putrescine in salt tolerance. Biochem Biophys Res Co 313:369–375
Watson MB, Malmberg RL (1996) Regulation of Arabidopsis thaliana L. Heynh arginine decarboxylase by potassium-deficiency stress. Plant Physiol 111:1077–1083
Williams GKJ (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol 167:766–778
Acknowledgments
The authors thank Professor Mark Johnson for the excellent facilities provided at the Structural Bioinformatics Laboratory of the Department of Biochemistry and Pharmacy, Åbo Akademi University. This work was supported by the Center for International Mobility (CIMO), Finland to S. Jantaro and the National Graduate School of Informational and Structural Biology, Svenska Kulturfonden and the Magnus Ehrnrooth Foundation to H. Kidron and by the Academy of Finland to T. Salminen and P. Mäenpää.
Author information
Authors and Affiliations
Corresponding author
Additional information
Saowarath Jantaro, Heidi Kidron, Tiina Salminen and Pirkko Mäenpää have contributed equally to this work
Rights and permissions
About this article
Cite this article
Jantaro, S., Kidron, H., Chesnel, D. et al. Structural modeling and environmental regulation of arginine decarboxylase in Synechocystis sp. PCC 6803. Arch Microbiol 184, 397–406 (2006). https://doi.org/10.1007/s00203-005-0064-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-005-0064-6