Abstract
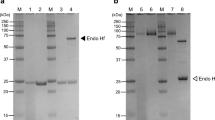
In spite of intensive exploitation of aspergilli for the industrial production of carbohydrases, little is known about hydrolytic enzymes of fungi from the section Cervini. Novel glycoside hydrolases Bgh12A and Xgh12B from Aspergillus cervinus represent examples of divergent activities within one enzyme family and belong to the GH12 phylogenetic subgroup I (endo-(1,4)-β-glucanases) and II (endo-xyloglucanases), respectively. The bgh12A and xgh12B genes were identified in the unsequenced genome of A. cervinus using primers designed for conservative regions of the corresponding subgroups and a genome walking approach. The recombinant enzymes were heterologously produced in Pichia pastoris, purified, and characterized. Bgh12A was an endo-(1,4)-β-glucanase (EC 3.2.1.4) hydrolyzing the unbranched soluble β-(1,4)-glucans and mixed linkage β-(1,3;1,4)-D-glucans. Bgh12A exhibited maximum activity on barley β-glucan (BBG), which amounted to 614 ± 30 U/mg of protein. The final products of BBG and lichenan hydrolysis were glucose, cellobiose, cellotriose, 4-O-β-laminaribiosyl-glucose, and a range of higher mixed-linkage gluco-oligosaccharides. In contrast, the activity of endo-xyloglucanase Xgh12B (EC 3.2.1.151) was restricted to xyloglucan, with 542 ± 39 U/mg protein. The enzyme cleaved the (1,4)-β-glycosidic bonds of the xyloglucan backbone at the unsubstituted glucose residues finally generating cellotetraose-based hepta-, octa, and nona-oligosaccharides. Bgh12A and Xgh12B had maximal activity at 55 °C, pH 5.0. At these conditions, the half-time of Xgh12B inactivation was 158 min, whereas the half-life of Bgh12A was 5 min. Recombinant P. pastoris strains produced up to 106 U/L of the target enzymes with at least 75% of recombinant protein in the total extracellular proteins. The Bgh12A and Xgh12B sequences show 43% identity. Strict differences in substrate specificity of Bgh12A and Xgh12B were in congruence with the presence of subgroup-specific structural loops and substrate-binding aromatic residues in the catalytic cleft of the enzymes. Individual composition of aromatic residues in the catalytic cleft defined variability in substrate selectivity within GH12 subgroups I and II.





Similar content being viewed by others
References
Angelov A, Pham VTT, Übelacker M, Brady S, Leis B, Pill N, Brolle J, Mechelke M, Moerch M, Henrissat B, Liebl W (2017) A metagenome-derived thermostable β-glucanase with an unusual module architecture which defines the new glycoside hydrolase family GH148. Sci Rep 7(1):17306. https://doi.org/10.1038/s41598-017-16839-8
Bauer S, Vasu P, Persson S, Mort AJ, Somerville CR (2006) Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. PNAS 103(30):11417–11422. https://doi.org/10.1073/pnas.0604632103
Berezina OV, Herlet J, Rykov SV, Kornberger P, Zavyalov A, Kozlov D, Sakhibgaraeva L, Krestyanova I, Schwarz WH, Zverlov VV, Liebl W, Yarotsky SV (2017) Thermostable multifunctional GH74 xyloglucanase from Myceliophthora thermophila: high-level expression in Pichia pastoris and characterization of the recombinant protein. Appl Microbiol Biotechnol 101(14):5653–5666. https://doi.org/10.1007/s00253-017-8297-2
Birsan C, Johnson P, Joshi M, MacLeod A, McIntosh L, Monem V, Nitz M, Rose DR, Tull D, Wakarchuck WW, Wang Q, Warren RA, White A, Withers SG (1998) Mechanisms of cellulases and xylanases. Biochem Soc Trans 26(2):156–160
Bukhtojarov FE, Ustinov BB, Salanovich TN, Antonov AI, Gusakov AV, Okunev ON, Sinitsyn AP (2004) Cellulase complex of the fungus Chrysosporium lucknowense: isolation and characterization of endoglucanases and cellobiohydrolases. Biochemistry (Mosc) 69:542–551
Calzado F, Prates ET, Gonçalves TA, Rubio MV, Zubieta MP, Squina FM, Skaf MS, Damásio AR (2016) Molecular basis of substrate recognition and specificity revealed in family 12 glycoside hydrolases. Biotechnol Bioeng 113(12):2577–2586. https://doi.org/10.1002/bit.26036
Christensen M, Fennell DI, Backus MP (1964) Aspergillus kanagawaensis and related species in Wisconsin forest soils. Mycologia 56(3):354–362. https://doi.org/10.2307/3756677
Dalboge H, Andersen LN, Kofod LV, Kauppinen S, Christgau S (1994) An enzyme with endoglucanase activity Patent WO 94/14953
Damasio AR, Ribeiro LF, Ribeiro LF, Furtado GP, Segato F, Almeida FB, Crivellari AC, Buckeridge MS, Souza TA, Murakami MT, Ward RJ, Prade RA, Polizeli ML (2012) Functional characterization and oligomerization of a recombinant xyloglucan-specific endo-β-1,4-glucanase (GH12) from Aspergillus niveus. Biochim Biophys Acta 1824(3):461–467. https://doi.org/10.1016/j.bbapap.2011.12.005
Damasio ARL, Rubio MV, Oliveira LC, Segato F, Dias BA, Citadini AP, Paixao DA, Fabio M, Squina FM (2014) Understanding the function of conserved variations in the catalytic loops of fungal glycoside hydrolase family 12. Biotechnol Bioeng 111(8):1494–1505. https://doi.org/10.1002/bit.25209
di Menna ME, Sayer ST, Barratt BIP, Bell NL, Ferguson CM, Townsend RJ (2007) Biodiversity of indigenous tussock grassland sites in Otago, Canterbury and the central North Island. V Penicillia and aspergilli. J Roy Soc New Zeal 37(3):131–137. https://doi.org/10.1080/03014220709510541
Dionex (1998) Technical note TN21: optimal settings for pulsed amperometric detection of carbohydrates using the Dionex ED40 electrochemical detector. Dionex, Sunnyvale
Forouchi E, Gunn DJ (1983) Some effects of metal ions on the estimation of reducing sugars in biological media. Biotechnol Bioeng 25(7):1905–1911. https://doi.org/10.1002/bit.260250717
Fry SC, York WS, Albersheim P, Darvill A, Hayashi T, Joseleau JP, Kato Y, Lorences EP, Maclachlan GA, McNeil M, Mort AJ, Reid JSG, Seitz HU, Selvendran RR, Voragen AGJ, White AR (1993) An unambiguous nomenclature for xyloglucan-derived oligosaccharides. Physiol Plant 89(1):1–3. https://doi.org/10.1111/j.1399-3054.1993.tb01778.x
Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, Kapitonov V, Jurka J, Scazzocchio C, Farman M, Butler J, Purcell S, Harris S, Braus GH, Draht O, Busch S, D'Enfert C, Bouchier C, Goldman GH, Bell-Pedersen D, Griffiths-Jones S, Doonan JH, Yu J, Vienken K, Pain A, Freitag M, Selker EU, Archer DB, Peñalva MA, Oakley BR, Momany M, Tanaka T, Kumagai T, Asai K, Machida M, Nierman WC, Denning DW, Caddick M, Hynes M, Paoletti M, Fischer R, Miller B, Dyer P, Sachs MS, Osmani SA, Birren BW (2005) Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438(7071):1105–1115. https://doi.org/10.1038/nature04341
Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA (2007) The current status of species recognition and identification in Aspergillus. Stud Mycol 59:1–10. https://doi.org/10.3114/sim.2007.59.01
Green MR, Sambrook J (2012) Molecular cloning: a laboratory manual, 4th edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Grishutin SG, Gusakov AV, Dzedzyulya EI, Sinitsyn AP (2006) A lichenase-like family 12 endo-(1-4)-β-glucanase from Aspergillus japonicus: study of the substrate specificity and mode of action on β-glucans in comparison with other glycoside hydrolases. Carbohydr Res 341(2):218–229. https://doi.org/10.1016/j.carres.2005.11.011
Houbraken J, Samson R (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70(1):1–51. https://doi.org/10.3114/sim.2011.70.01
Houbraken J, de Vries RP, Samson RA (2014) Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol 86:199–249. https://doi.org/10.1016/B978-0-12-800262-9.00004-4
Huang JW, Liu W, Lai HL, Cheng YS, Zheng Y, Li Q, Sun H, Kuo CJ, Guo RT, Chen CC (2016) Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50. Biochem Biophys Res Commun 478:565–572. https://doi.org/10.1016/j.bbrc.2016.07.101
Iakiviak M, Mackie RI, Cann IKO (2011) Functional analyses of multiple lichenin-degrading enzymes from the rumen bacterium Ruminococcus albus 8. Appl Environ Microbiol 2011(7):7541–7550. https://doi.org/10.1128/AEM.06088-11
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8(3):275–282
Kanda T, Yatomi H, Makishima S, Amano Y, Nisizawa K (1989) Substrate specificities of exo- and endo-type cellulases in the hydrolysis of β-(1,3)- and β-(1,4)-mixed D-glucans. J Biochem 105:127–132. https://doi.org/10.1093/oxfordjournals.jbchem.a122606
Khademi S, Zhang D, Swanson SM, Wartenberg A, Witte K, Meyer EF (2002) Determination of the structure of an endoglucanase from Aspergillus niger and its mode of inhibition by palladium chloride. Acta Cryst D58:660–667. https://doi.org/10.1107/S0907444902003360
Kocsubé S, Perrone G, Magistà D, Houbraken J, Varga J, Szigeti G (2016) Aspergillus is monophyletic: evidence from multiple gene phylogenies and extrolites profiles. Stud Mycol 85:199–213. https://doi.org/10.1016/j.simyco.2016.11.006
Krestyanova IN, Sakhibgaraeva LF, Berezina OV, Rykov SV, Zavyalov AV, Zverlov VV, Yarotsky SV (2016) Characteristics of fungal strains producing thermostable xyloglucanases from the Russian National Collection of industrial microorganisms. Mol Genet Microbiol Virol 31(3):149–155. https://doi.org/10.3103/S089141681603006X
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. https://doi.org/10.1093/molbev/msw054
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lee HS, Zhang Y (2012) BSP-SLIM: a blind low-resolution ligand-protein docking approach using theoretically predicted protein structures. Proteins 80(1):93–110. https://doi.org/10.1002/prot.23165
Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. https://doi.org/10.1093/nar/gkt1178
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193(1):265–275
Macarron R, Acebal C, Castillon MP, Dominguez JM, de la Mata I, Pettersson G, Tomme P, Claeyssens M (1993) Mode of action of endoglucanase III from Trichoderma reesei. Biochem J 289(Pt 3):867–873. https://doi.org/10.1042/bj2890867
Master ER, Zheng Y, Storms R, Tsang A, Powlowski J (2008) A xyloglucan-specific family 12 glycosyl hydrolase from Aspergillus niger: recombinant expression, purification and characterization. Biochem J 411(1):161–170. https://doi.org/10.1042/BJ20070819
Matz M, Shagin D, Bogdanova E, Britanova O, Lukyanov S, Diatchenko L, Chenchik A (1999) Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res 27(6):1558–1560. https://doi.org/10.1093/nar/27.6.1558
Mechelke M, Herlet J, Benz JP, Schwarz WH, Zverlov VV, Liebl W, Kornberger P (2017) HPAEC-PAD for oligosaccharide analysis – novel insights into analyte sensitivity and response stability. Anal Bioanal Chem 409(30):7169–7181. https://doi.org/10.1007/s00216-017-0678-y
Meng DD, Liu X, Dong S, Wang YF, Ma XQ, Zhou HX, Wang XQ, Yao LS, Feng YG, Li FL (2017) Structural insights into the substrate specificity of a glycoside hydrolase family 5 lichenase from Caldicellulosiruptor sp. F32. Biochem J 474(20):3373–3389. https://doi.org/10.1042/BCJ20170328
Nakazawa H, Okada K, Kobayashi R, Kubota T, Onodera T, Ochiai N, Omata N, Ogasawara W, Okada H, Morikawa Y (2008) Characterization of the catalytic domains of Trichoderma reesei endoglucanase I, II, and III, expressed in Escherichia coli. Appl Microbiol Biotechnol 81:681–689. https://doi.org/10.1007/s00253-008-1667-z
Neill JC (1939) The mould fungi of New Zealand. II. The genus Aspergillus. Trans Proc Royal Soc N Z 69:237–264
Nielsen RI (2002) Microbial xyloglucan endotransglycosylase (XET). Patent US 6448056 B1
Nishinari K, Takemasa M, Zhang H, Takahashi R (2007) Storage plant polysaccharides: xyloglucans, galactomannans, glucomannans. In: Kamerling JP, Boons G-J, Lee YC, Suzuki A, Taniguchi N, Voragen AGJ (eds) Comprehensive glycoscience. From chemistry to systems biology. chapter 2.19. Elsevier Ltd, Amsterdam, pp 613-652. doi:https://doi.org/10.1016/B978-044451967-2/00146-X
Noss I, Doekes G, Thorne PS, Heederik DJJ, Wouters IM (2013) Comparison of the potency of a variety of β-glucans to induce cytokine production in human whole blood. Innate Immun 19(1):10–19. https://doi.org/10.1177/1753425912447129
Ooi T, Shinmyo A, Okada H, Hara S, Ikenaka T, Murao S, Arai M (1990) Cloning and sequence analysis of a cDNA for cellulase (FI-CMCase) from Aspergillus aculeatus. Curr Genet 18(3):217–222
Park YW, Baba K, Furuta Y, Iida I, Sameshima K, Arai M, Hayashi T (2004) Enhancement of growth and cellulose accumulation by overexpression of xyloglucanase in poplar. FEBS Lett 564(1-2):183–187. https://doi.org/10.1016/S0014-5793(04)00346-1
Peat S, Whelan WJ, Roberts JG (1957) The structure of lichenin. J Chem Soc:3916–3924. https://doi.org/10.1039/JR9570003916
Perlin AS, Suzuki S (1962) The structure of lichenin: selective enzymolysis studies. Can J Chem 40(1):50–56. https://doi.org/10.1139/v62-009
Prates ET, Stankovic I, Silveira RL, Liberato MV, Henrique-Silva F, Pereira N, Polikarpov I, Skaf MS (2013) X-ray structure and molecular dynamics simulations of endoglucanase 3 from Trichoderma harzianum: structural organization and substrate recognition by endoglucanases that lack cellulose binding module. PLoS One 8(3):e59069–e59069. https://doi.org/10.1371/journal.pone.0059069
Rawat R, Kumar S, Chadha BS, Kumar D, Oberoi HS (2015) An acidothermophilic functionally active novel GH12 family endoglucanase from Aspergillus niger HO: purification, characterization and molecular interaction studies. Antonie Van Leeuwenhoek 107(1):103–117. https://doi.org/10.1007/s10482-014-0308-z
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sakamoto S, Tamura G, Ito K, Ishikawa T, Iwano K, Nishiya N (1995) Cloning and sequencing of cellulase cDNA from Aspergillus kawachii and its expression in Saccharomyces cerevisiae. Curr Genet 27(5):435–439
Samson RA, Visagie CM, Houbraken J, Hong S-B, Hubka V, Klaassen CHW, Perrone G, Seifert KA, Susca A, Tanney JB, Varga J, Kocsubé S, Szigeti G, Yaguchi T, Frisvad JC (2014) Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol 78:141–173. https://doi.org/10.1016/j.simyco.2014.07.004
Sandgren M, Shaw A, Ropp TH, Wu S, Bott R, Cameron AD, Stahlberg J, Mitchinson C, Jones TA (2001) The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 A resolution. J Mol Biol 308:295–310. https://doi.org/10.1006/jmbi.2001.4583
Sandgren M, Gualfetti PJ, Shaw A, Gross LS, Saldajeno M, Day AG, Jones TA, Mitchinson C (2003) Comparison of family 12 glycoside hydrolases and recruited substitutions important for thermal stability. Protein Sci 12(4):848–860. https://doi.org/10.1110/ps.0237703
Sandgren M, Berglund GI, Shaw A, Stahlberg J, Kenne L, Desmet T, Mitchinson C (2004) Crystal complex structures reveal how substrate is bound in the -4 to the +2 binding sites of Humicola grisea Cel12A. J Mol Biol 342(5):1505–1517. https://doi.org/10.1016/j.jmb.2004.07.098
Sandgren M, Ståhlberg J, Mitchinson C (2005) Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes. Prog Biophys Mol Biol 89(3):246–291. https://doi.org/10.1016/j.pbiomolbio.2004.11.002
Segato F, Dias B, Berto GL, de Oliveira DM, De Souza FHM, Citadini AP, Murakami MT, Damásio ARL, Squina FM, Polikarpov I (2017) Cloning, heterologous expression and biochemical characterization of a non-specific endoglucanase family 12 from Aspergillus terreus NIH2624. Biochim Biophys Acta 1865(4):395–403. https://doi.org/10.1016/j.bbapap.2017.01.003
Shi H, Yin X, Wu M, Tang C, Zhang H, Li J (2012) Cloning and bioinformatics analysis of an endoglucanase gene (Aucel12A) from Aspergillus usamii and its functional expression in Pichia pastoris. J Ind Microbiol Biotechnol 39(2):347–357. https://doi.org/10.1007/s10295-011-1039-z
Sinegani AAS, Emtiazi G (2006) The relative effects of some elements on the DNS method in cellulase assay. JASEM 10(3):93–96. dx.doi.org. https://doi.org/10.4314/jasem.v10i3.17326
Synytsya A, Novak M (2014) Structural analysis of glucans. Ann Transl Med 2(2):17. https://doi.org/10.3978/j.issn.2305-5839.2014.02.07
Takeda T, Nakano Y, Takahashi M, Sakamoto Y, Konno N (2013) Polysaccharide-inducible endoglucanases from Lentinula edodes exhibit a preferential hydrolysis of 1,3-1,4-β-glucan and xyloglucan. J Agric Food Chem 61(31):7591–7598. https://doi.org/10.1021/jf401543m
Tsang CC, Tang JYM, Lau SKP, Woo PCY (2018) Taxonomy and evolution of Aspergillus, Penicillium and Talaromyces in the omics era – past, present and future. Comput Struct Biotechnol J 16:197–210. https://doi.org/10.1016/j.csbj.2018.05.003
Vincken J-P, Beldman G, Voragen AGJ (1997) Substrate specificity of endoglucanases: what determines xyloglucanase activity? Carbohydr Res 298:299–310
Wang Z, Ye S, Li J, Zheng B, Bao M, Ning G (2011) Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol 11:109. https://doi.org/10.1186/1472-6750-11-109
Wood TM, Bhat KM (1988) Methods for measuring cellulase activities. Methods Enzymol 160:87–117. https://doi.org/10.1016/0076-6879(88)60109-1
Yang H, Shi P, Liu Y, Xia W, Wang X, Cao H, Ma R, Luo H, Bai Y, Yao B (2017) Loop 3 of fungal endoglucanases of glycoside hydrolase family 12 modulates catalytic efficiency. Appl Environ Microbiol 83(6). doi:https://doi.org/10.1128/AEM.03123-16
Yoshizawa T, Shimizu T, Hirano H, Sato M, Hashimoto H (2012) Structural basis for inhibition of xyloglucan-specific endo-β-1,4-glucanase (XEG) by XEG-protein inhibitor. J Biol Chem 287:18710–18716. https://doi.org/10.1074/jbc.M112.350520
Zavyalov AV, Rykov SV, Lunina NA, Sushkova VI, Yarotsky SV, Berezina OV (2018) Plant polysaccharide xyloglucan and enzymes that hydrolyze it. Chem Plant Raw Mater 4:43–61. https://doi.org/10.14258/jcprm.2018043926
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166. https://doi.org/10.1016/B978-1-4832-2734-4.50017-6
Funding
This study was funded by the Ministry of Education and Science of the Russian Federation (grant number 14.628.21.0001) and the German Federal Ministry of Education and Research (grant number FKZ: 031A556).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The article does not contain any studies with human participants or animals performed by any of the authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 1224 kb)
Rights and permissions
About this article
Cite this article
Rykov, S.V., Kornberger, P., Herlet, J. et al. Novel endo-(1,4)-β-glucanase Bgh12A and xyloglucanase Xgh12B from Aspergillus cervinus belong to GH12 subgroup I and II, respectively. Appl Microbiol Biotechnol 103, 7553–7566 (2019). https://doi.org/10.1007/s00253-019-10006-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-019-10006-x