Abstract
Integral membrane transporters of the Mycobacterial Membrane Protein Large (MmpL) family and their interactome play important roles in the synthesis and export of mycobacterial outer membrane lipids. Despite the current interest in the mycolic acid transporter, MmpL3, from the perspective of drug discovery, the nature and biological significance of its interactome remain largely unknown. We here report on a genome-wide screening by two-hybrid system for MmpL3 binding partners. While a surprisingly low number of proteins involved in mycolic acid biosynthesis was found to interact with MmpL3, numerous enzymes and transporters participating in the biogenesis of peptidoglycan, arabinogalactan and lipoglycans, and the cell division regulatory protein, CrgA, were identified among the hits. Surface plasmon resonance and co-immunoprecipitation independently confirmed physical interactions for three proteins in vitro and/or in vivo. Results are in line with the focal localization of MmpL3 at the poles and septum of actively-growing bacilli where the synthesis of all major constituents of the cell wall core are known to occur, and are further suggestive of a role for MmpL3 in the coordination of new cell wall deposition during cell septation and elongation. This novel aspect of the physiology of MmpL3 may contribute to the extreme vulnerability and high therapeutic potential of this transporter.
Similar content being viewed by others
Introduction
Mycolic acids are essential building blocks of the outer membrane of all mycobacteria. The importance of drugs targeting their biosynthesis and export is illustrated by the therapeutic efficacy of such anti-mycobacterial agents as isoniazid, ethionamide, thiacetazone and isoxyl, and a number of recently discovered small molecule inhibitors reported to inhibit the integral membrane mycolic acid transporter, MmpL31,2. Studies from our group and others have established that MmpL3, a member of the Resistance, Nodulation and Division (RND) superfamily, is required for the translocation of mycolic acids in the form of trehalose monomycolates (TMM) from the cytoplasm to the periplasm or outer membrane where this glycolipid can then serve as a mycolic acid donor for the enzymes catalyzing their transesterification to arabinogalactan or extracellular TMM yielding trehalose dimycolates (TDM) [Fig. 1]3,4,5.
The M. tuberculosis mycolic acid biosynthetic pathway. The C48–C54 meromycolate chain is biosynthesized by FAS-II through the processive addition of multiple malonate units onto C16–C26 precursors generated by FAS-I. The initial substrates of FAS-II are thus medium length keto-acyl-ACP resulting from the condensation by the M. tuberculosis FabH protein of the acyl-CoA products of FAS-I with malonyl-ACP. After reduction by the β-keto-acyl-ACP reductase MabA, dehydration by the (3R)-hydroxyacyl dehydratases HadAB and HadBC, and reduction by the enoyl-CoA reductase InhA, either the β-ketoacyl-ACP synthase KasA or KasB catalyzes the condensation of the resulting product with malonyl-ACP units, thereby initiating the next round of elongation. The products of FAS-II may undergo further elongation and functional modifications of the meromycolic acid chain, catalyzed in part by S-adenosyl methionine-dependent methyltransferases (MmaA1, MmaA2, MmaA3, MmaA4, CmaA1, CmaA2, PcaA, UmaA), prior to the Pks13/FadD32-mediated condensation of the activated α-branch with the meromycolic acid chain to yield the full-size mycolic β-ketoester. Pks13 transfers mycolic β-ketoester to trehalose prior to reduction by the CmrA reductase yielding mature TMM. TmaT acetylates the mycolic acid in TMM prior to its export by MmpL3 and other potential components of a translocation machinery spanning the mycobacterial cell envelope. The mycolyltransferases encoded by fbpA, fbpB and fbpC catalyze the transfer of mycolic acids from TMM to arabinogalactan (AG), or to another TMM molecule generating TDM69. Proteins in red font were tested individually for interaction with MmpL3 by BATCH two-hybrid system.
Interestingly, multiple cell envelope-related biosynthetic pathways (e.g., sulfolipids, phthiocerol dimycocerosates, polyacyltrehaloses, glycopeptidolipids, siderophores, etc.) in mycobacteria and closely related Actinomycetes rely on related MmpL (mycobacterial membrane proteins, large) proteins for the translocation of their products to their final extra-cytoplasmic location6,7. MmpL proteins typically interact with other enzymes and transporters of the same biosynthetic pathway and these physical interactions are required for efficient coupling of synthesis and export6,7,8. Although earlier work has begun to shed light on the nature and functional relevance of physical interactions between mycolic acid biosynthetic enzymes9,10,11,12, it has thus far only focused on known cytosolic enzymes and has not integrated any aspects of the translocation machinery. As a result, nothing is presently known of the spatial and temporal coupling of mycolic acid biosynthesis and export.
Given the challenge represented by the translocation of such high molecular weight, hydrophobic, constituents as mycolic acids across the different layers of the mycobacterial cell envelope, the existence of a molecular scaffold providing continuity between the cytoplasm, periplasm and the surface of mycobacteria also seems likely. This assumption is further supported by our single-particle electron microscopy studies showing that the length of MmpL3 (100 Å of which an estimated 35 Å extends in the periplasmic space) is not compatible with direct substrate delivery to the outer membrane (the thickness of the mycobacterial cell envelope is in the range of ∼35–40 nm)13,14,15. The first evidence of the existence of accessory proteins required for TMM export was obtained very recently using a co-purification strategy to identify proteins interacting with MmpL3 in Mycobacterium smegmatis. One plasma membrane-anchored protein named TtfA was shown to be required for TMM export whereas another protein co-eluting with MmpL3 was proposed to play a role in the stabilization of the MmpL3-TtfA complex under stress conditions16. Whether additional periplasmic adapters, outer membrane proteins and, perhaps, inner membrane transporters work with MmpL3 to bring TMM to the cell surface, similarly to the situation with prototypical HAE1 RND efflux pumps17 remains to be determined.
Another unique aspect of the biology of mycobacteria and other bacteria of the order Actinomycetales is the fact that insertion of newly synthesized peptidoglycan during elongation occurs at the cell poles rather than along the lateral cell wall as in other rod-shaped bacteria such as Escherichia coli and Bacillus subtilis18,19. Bacilli division and elongation without compromising the integrity of their existing multilayered cell wall implies spatiotemporal coordination of biosynthetic and export activities at the sites of septal and polar elongation, a process apparently facilitated in mycobacteria by the uneven distribution of the peptidoglycan, arabinogalactan, mycolic acid and phospho(glyco)lipid synthetic complexes in the inner membrane, and their enrichment in the polar and subpolar regions of actively elongating cells20,21,22,23,24. Despite these advances, much remains to be done in understanding the molecular mechanisms coordinating new cell wall deposition with cell septation/elongation, and characterizing the molecular interactions and dynamic features of the individual cell envelope biosynthetic pathways involved. The central involvement of MmpL3 in mycolic acid translocation, the localization of this transporter at the pole and septum of actively-growing cells20,25 where mycolic acid transfer to their outer membrane acceptors has been shown to occur26,27,28,29, and its ability to directly or indirectly interact with the elongation regulator Wag31 (aka DivIVA)20 led us to hypothesize that MmpL3 may be at the center of a protein scaffold coordinating new mycolic acid and, perhaps, other cell envelope constituents deposition during cell elongation and division.
The present interactome study was thus undertaken with three main aims. The first one was to determine whether MmpL3 acts as a scaffold for a multiprotein complex coupling mycolic acid synthesis and export. The second one was to identify putative additional components of the mycolic acid translocation machinery from M. tuberculosis. The third one was to begin delineating the other key events at the intersection of cell wall biogenesis, cell elongation and cell division in which MmpL3 may be involved. The results of these studies point to a complex crosstalk between mycolic acid, lipid and polysaccharide biosynthesis, and cell elongation and division in mycobacteria.
Results
Identification of MmpL3 binding partners using a bacterial two-hybrid system
We resorted to the E. coli adenylate cyclase two-hybrid (BACTH) system as our primary approach to identify M. tuberculosis proteins interacting with MmpL3. This system exploits the fact that the catalytic domain of the adenylate cyclase (CyaA) from Bordetella pertussis consists of two complementary fragments, T25 and T18, that are not active when physically separated but whose functional complementation when fused to interacting polypeptides results in cyclic AMP synthesis in an E. coli cya mutant30. cAMP produced by the reconstituted chimeric enzyme activates a β-galactosidase reporter allowing positive interactions to be easily identified on indicator agar media. Importantly, since this system relies on a signaling cascade that utilizes a diffusible regulatory molecule, it is suitable to detect interactions between cytoplasmic, as well as transmembrane and membrane-associated proteins31,32.
MmpL3 fusion proteins harboring C-terminal or N-terminal T18 and T25 domains were generated as “baits” and systematically screened for pairwise interactions with four major sets of similarly fused proteins. Functional complementation efficiency between the T18 and T25 domains was quantified by measuring β-galactosidase activity. The first “unbiased” set, which was initially screened as pools of 96 baits against MmpL3, contained the entire M. tuberculosis Gateway® Clone Set from BEI Resources (NR-19274) consisting of 3,724 unique M. tuberculosis H37Rv and CDC1551 open reading frames (ORFs). Since the M. tuberculosis Gateway® Clone Set is missing a number of M. tuberculosis H37Rv ORFs, and because we also wanted to screen a number of target proteins individually against MmpL3, T18 and T25 C- and N-terminally-fused proteins were generated for the sets of test proteins whose description follows. One of these sets is represented on Fig. 1 and consists of all of the proteins known to be involved in mycolic acid biosynthesis. Given the fact that genes involved in the production and MmpL-dependent export of lipids tend to cluster in the genomes of mycobacteria6, another set of candidate interactors was further generated which consisted of all of the conserved genes clustering with mmpL3 in M. tuberculosis and other mycobacterial genomes [Fig. 2]. Finally, fusions were prepared for candidate proteins that had either been proposed to interact with MmpL3 (e.g., Wag31)20 or were known to participate in lipid export in other MmpL-dependent pathways (e.g., Sap [Rv3821], the Sap-like protein Rv1517, and Rv0585c)6,7.
In toto, this in vivo screen yielded 19 hits that reproducibly interacted with MmpL3 by BACTH two-hybrid system. These hits, their proposed localization in the cell and putative function are presented in Table 1. The results of the β-galactosidase activity assays are presented in Figs 3 and S1. Overall, the results which are detailed below highlighted a broad network of proteins participating in cell envelope biogenesis, cell elongation/division and as yet unknown functions with which MmpL3 appears to interact in vivo.
Quantification of in vivo interactions between full-size or C-terminal truncated MmpL3 and protein partners. MmpL3 (full-size) and MmpL31–744 fusion proteins harboring C-terminal or N-terminal T18 and T25 domains were generated as “baits” and systematically compared for pairwise interactions with the binding partners listed in Table 1. Pairwise co-transformants developing a blue color in the BATCH screen were grown in LB broth, and the cultures processed for β-galactosidase activity as described in Methods. The values presented are the mean activities (relative units) ± standard error from measurements performed on three independent E. coli transformants.
Interaction of MmpL3 with components of the mycolic acid biosynthetic machinery and products of syntenic genes
Consistent with our earlier finding that the full-size MmpL3 protein functions as a homotrimer15, MmpL3 interacted with itself [Fig. 3]. Of the known mycolic acid biosynthetic enzymes that might have interacted with MmpL3 in the cytoplasm or in the membrane itself, none of the enzymes involved in the synthesis of meromycolate precursors (e.g., FAS-II enzymes), functionalization of the meromycolate chain (S-adenosyl-methionine methyltransferases) or final assembling of the meromycolate with the alpha-branch (Pks13, FadD32) [Fig. 1] were identified as MmpL3 binding partners under the assay conditions used herein [Fig. S1]. In fact, the only interactor identified within this pathway was TmaT, an acyltransferase responsible for the acetylation of mycolic acids in TMM whose activity appears to be a requisite for TMM export and cell viability in mycobacteria15,33. TmaT is thought to catalyze the last modification of TMM prior to export by MmpL3. tmaT clusters with mmpL3 in the same genomic region, as do a number of other binding partners identified in this screen (MmpL11; Rv0204c; Rv0207c; Rv0227c; AftD). Rv0207c, a Mycobacterium-conserved gene of unknown function, maps adjacent to mmpL3. Rv0204c encodes a potential inner membrane transporter with homology to MprF translocases. Multiple Peptide Resistance Factor (MprF) proteins represent a highly conserved protein family responsible for the synthesis and translocation of aminoacyl phospholipids in a variety of bacteria34. Rv0204c, however, is unusual in that it is devoid of the biosynthetic domain of MprF proteins and only harbors the translocase domain. MmpL11 (Rv0202c) is involved in the translocation of the mycolic acid-containing lipids, monomeromycolyl diacylglycerol and mycolate wax ester35 and, together with MmpL3, was proposed to participate in heme-iron uptake on the basis of the ability of the periplasmic domains of these two RND transporters to bind heme36,37. While the two transporters only showed weak interaction between them [Fig. 3], they both were identified as binding partners of the MprF-like translocase Rv0204c [Figs 3 and 4] suggestive of some form of coupling of the two MmpL-dependent pathways. Although Rv0204c is not required for growth, the disruption of this gene has been reported to attenuate M. tuberculosis virulence38,39. Rv0227c was recently shown to serve a critical, albeit incompletely defined, function in the elongation of lipomannan (LM) and lipoarabinomannan (LAM) in Corynebacterium glutamicum40, while AftD (Rv0236c) plays an essential role in the arabinosylation of LAM and arabinogalactan in mycobacteria41,42.
Quantification of in vivo interactions between MmpL3 binding partners. CrgA, Wag31, TmaT, AftD, Rv0227c, Rv0204c and MmpL11 fusion proteins harboring C-terminal or N-terminal T18 and T25 domains were generated as “baits” and tested for pairwise interactions with other MmpL3 binding partners. Pairwise co-transformants developing a blue color in the BATCH screen were grown in LB broth, and the cultures processed for β-galactosidase activity as in Fig. 3.
Lack of periplasmic interactions between MmpL3 and the FbpA, FbpB and FbpC mycolyltransferases
Three mycolyltransferases in M. tuberculosis - FbpA (Rv3804c), FbpB (Rv1886c) and FbpC (Rv0129c) - catalyze, on the periplasmic side of the plasma membrane, the transfer of mycolic acids from TMM to their cell envelope acceptors [Fig. 1]. Since the periplasmic location of these enzymes precluded the use of the BACTH system to detect potential interactions with MmpL3, Surface Plasmon Resonance (SPR) was used instead. To this end, the MmpL3 protein from M. tuberculosis was produced in a M. smegmatis knock-out mutant devoid of its endogenous mmpL3 gene and purified using a combination of nickel affinity and ion-exchange chromatography [Fig. S2]. The purified MmpL3 protein was then immobilized onto the surface of CM5 chips as described under Methods, and increasing concentrations of the analyte proteins were injected over the surface. SPR assays with the purified native FbpA, FbpB and FbpC proteins failed to reveal any interaction with MmpL3 [Fig. S3]. Similarly, testing of purified Pks13 protein by SPR confirmed the lack of interaction between MmpL3 and this polyketide synthase [Fig. S3].
MmpL3 binding partners with other known or proposed functions in cell envelope biogenesis
Interestingly, the BACTH screening identified a number of other MmpL3-binding partners outside the mmpL3 cluster whose demonstrated or proposed function, or clustering with cell envelope-related genes in the M. tuberculosis genome, are suggestive of their participation in cell envelope biogenesis. Foremost among these are three highly conserved proteins involved, or thought to be involved, in peptidoglycan synthesis and export: Rv1337, Rv3909 and Rv3910 (MviN) [Fig. 3]. MviN is required for peptidoglycan precursor export or polymerization; by analogy with Gram-negative MviN proteins, it was proposed to act as a lipid II flippase43. Rv3909 was predicted by bioinformatics approaches to be an outer membrane protein44 although experimental proof for this assumption is currently lacking. The organization of the Rv3909 and mviN loci in the M. tuberculosis genome suggests that the two genes are probably co-transcribed. Rv1337 is a conserved rhomboid-like protein of unknown function whose encoding gene clusters with the essential peptidoglycan biosynthetic gene murI in all mycobacterial genomes45. Disruption of this gene in M. smegmatis was found to alter the ability of bacilli to form biofilms and their susceptibility to hydrophobic antibiotics suggestive of changes in cell surface properties and cell envelope permeability46.
Other interactors potentially involved in cell envelope synthesis include Rv0625c, a protein of unknown function whose encoding gene clusters with a number of arabinogalactan (galTa, galTb) and mycolic acid biosynthetic genes (hadA, hadB, hadC). Rv1457c, an inner membrane component of an ABC-transporter whose genetic association with a mannosyltransferase involved in LM/LAM synthesis (Rv1459c/MptB) is suggestive of its involvement in the export of lipoglycans47. Rv2169c maps in a region of the M. tuberculosis genome rich in recently characterized cell division regulator genes (Rv2151, Rv2164)48 as well as peptidoglycan, LM, LAM and phospholipid biosynthetic genes (Rv2163c/pbpB; Rv2174/mptA, Rv2181, Rv2182c)49. Rv3483c is adjacent to the cell wall ligase gene cpsA2 (Rv3484) involved in the covalent attachment of arabinogalactan with peptidoglycan50, and the ortholog of this protein in M. marinum is apparently secreted by the ESX-1 secretion system51. All of these proteins and other interactors of as yet unknown function presented in Table 1 (Rv3064c, Rv3271c, LprC, LppT) are predicted to be periplasmic, integral membrane or outer membrane proteins and may participate in the export of TMM or that of other as yet unknown cell envelope constituents. We note that the finding of lipoproteins and other potentially exported proteins in our screen was unexpected as the BACTH system is not designed to reveal extra-cytoplasmic interactions. These results may indicate false-positives. Alternatively, the improper export or insertion in the plasma membrane of some of the test proteins and MmpL3 fusions produced in E. coli may inadvertently have led extra-cytoplasmic interactors of MmpL3 to interact with periplasmic regions of this transporter in the cytoplasm of E. coli.
Interaction of MmpL3 with proteins involved in cell elongation and septation
An earlier interactome study identified MmpL3 among proteins co-immunoprecipitating with Wag3120, an elongation specific regulatory factor involved in the coordination of cell envelope biogenesis at the older, faster growing, pole of mycobacteria21. While the BACTH assay failed to reveal any direct interaction between MmpL3 and Wag31 (data not shown), the screening of the M. tuberculosis genomic “prey” library from BEI Resources identified CrgA as an MmpL3 interactor [Fig. 3]. CrgA is a cell division regulatory protein that facilitates septum formation and ensures proper septal and polar peptidoglycan assembly by influencing the localization of penicillin-binding proteins and Wag3152,53. The identification of CrgA as a protein interacting with MmpL3 unveils a plausible mechanism through which MmpL3 may be recruited to the septum to drive mycolic acid deposition at this site during cell division25,29. Given that CrgA interacts with Wag31 [Fig. 4], it is possible that CrgA further contributes to the relocalization of MmpL3 to the old poles during cell elongation21,25,29.
Validation of MmpL3 interactions with CrgA, Rv0207c and AftD by surface plasmon resonance
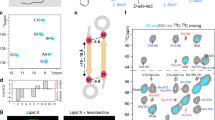
We next turned to an independent in vitro protein-protein interaction approach, namely SPR, to validate the results of some of our BACTH assays with a subset of proteins selected to encompass the variety of functions (mycolic acid biosynthesis, peptidoglycan arabinogalactan and lipoarabinomannan synthesis, cell elongation and division) and subcellular localizations (inner membrane, periplasm, cytoplasm) uncovered by the two-hybrid system screening. To this end, the MviN, AftD, LprC, CrgA and Rv0207c proteins were produced in E. coli and purified by a combination of ion exchange and metal affinity chromatographies as described under Methods [Fig. S2]. Attempts to produce and purify TmaT unfortunately failed, both in E. coli and M. smegmatis. The purified MmpL3 was then captured onto the surface of CM5 chips and interactions with the candidate proteins were analyzed as described for FbpA, FbpB, FbpC and Pks13, by injecting over the surface increasing concentrations of MviN, AftD, LprC, CrgA and Rv0207c. The results, which are presented in Fig. 5, indicated that CrgA, Rv0207c and AftD, but not LprC or MviN [Fig. S3], bind specifically and with high affinities to the immobilized MmpL3. The collected CrgA, AftD and Rv0207c sensorgrams were fit globally into different kinetic models to analyze the mechanism and affinity of the interactions. The goodness of obtained fits was comparable for different models, and a simple 1:1 binding model was used further to compare the affinities of the observed protein-protein interactions. For all three proteins, the dissociation constants (KD) for the interaction with MmpL3 were found to be in the mid nanomolar range [Fig. 5 and Table S1]. Thus, MmpL3 interacts directly and with comparable affinities with CrgA, AftD and Rv0207c. Failure to detect an interaction between LprC and MmpL3 supports our earlier assumption that possibly several, if not all, of the interactions revealed by BACTH between MmpL3 and exported proteins may in fact be false positives. The lack of detectable interaction between MviN and MmpL3 in the SPR assay may likewise either indicate a false positive or result from the improper folding or oligomerization of the recombinant form of MviN produced in E. coli.
Quantification of in vitro interactions between MmpL3 and binding partners by surface plasmon resonance. SPR was used to analyze kinetics of the purified MmpL3 interaction with various proteins. Binding sensorgrams were collected by injecting two-fold increasing concentrations ranging from 0.6 μM up to 10 μM of AftD (violet), Rv0207c (blue) and CrgA (green). Binding curves were fitted globally into a simple 1:1 model (red lines) and the dissociations constants are shown for each protein. No specific binding was detected for the Ag85 complex, Pks13, MviN and LprC [see Fig. S3].
MmpL3 interaction with CrgA in intact M. smegmatis cells
To determine whether MmpL3 interacted with CrgA in vivo in the context of intact M. smegmatis cells, we finally resorted to co-immunoprecipitation. To this end, a recombinant CrgA protein N-terminally-tagged with a FLAG epitope was expressed in the background of MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp from the integrative plasmid pFAX-crgA. Intact M. smegmatis cells co-expressing CrgA-FLAG and MmpL3tb-GFP tagged with hexahistidine at the C-terminal end were treated with the cross-linking agent dithiobis[succinimidyl propionate] (DSP) to cross-link protein complexes before breaking the cells. MmpL3tb was next purified from these cells by Ni-NTA affinity chromatography, and the presence of CrgA-FLAG in MmpL3-containing elution fractions was investigated by immunoblot using antibodies to the FLAG epitope. The elution fractions from M. smegmatis cells missing either the expression plasmid for CrgA-FLAG or pMVGH1-mmpL3tb-gfp were used as negative controls. As shown in Fig. 6, CrgA-FLAG co-eluted with MmpL3tb-GFP, yielding a FLAG-positive signal of the expected size for CrgA-FLAG (~12 kDa) upon reduction of protein complexes with dithiothreitol. In contrast, CrgA-FLAG was missing in the pulldown samples similarly prepared from M. smegmatis cells missing either pFAX-crgA or pMVGH1-mmpL3tb-gfp [Figs 6 and S4]. Thus, CrgA appears to interact with MmpL3tb in intact mycobacterial cells. Either due to low abundance or to untoward exposure of the FLAG epitope within protein complexes, CrgA-FLAG was not directly detected in the high molecular weight MmpL3-GFP complexes and was only visible upon DTT treatment [Fig. 6]. Furthermore, no CrgA was detected in the elution fraction when the crosslinker was omitted from the cell preparation (data not shown).
CrgA interacts with MmpL3 in intact mycobacterial cells. Left: In-gel fluorescence of DSP-treated, detergent-solubilized, MmpL3tb-protein complexes prepared from MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp + pFAX-crgA cells reveals the presence of high molecular weight MmpL3-GFP protein complexes in the elution fractions that are reduced upon addition of DTT. The expected size of MmpL3-GFP is ~126 KDa. Right: Immunoblot analysis of DSP-treated, detergent-solubilized, MmpL3tb-protein complexes prepared from Msmg + pFAX-crgA and MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp + pFAX-crgA cells. The immunoblot shows the presence of CrgA-FLAG in the elution fractions from MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp + pFAX-crgA cells but not in those from cells devoid of mmpL3tb-gfp expression plasmid. The expected size of CrgA-FLAG is ~12 KDa. *Denotes a non-specific M. smegmatis protein reacting with the anti-FLAG antibody. The full-length gel and blots are shown. Co-affinity purifications were performed twice on independent culture batches with the same results.
Impact of the C-terminal cytoplasmic domain of MmpL3 on protein interactions
The ability of MmpL3 to accumulate at the old pole and septa of actively growing cells was shown to be dependent on a region of the transporter located between the end of its tenth transmembrane segment (TMS) and the C-terminus25. To more precisely delineate the region of MmpL3 required for its dynamic localization in growing bacilli, M. smegmatis recombinant strains were generated of which the endogenous mmpL3 gene was entirely deleted and replaced by one of two different, ectopically expressed, rescue copies of the mmpL3 gene from M. tuberculosis. One rescue copy corresponded to the full-size mmpL3 gene fused at its 3′ end to gfp (yielding MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp). The second rescue copy corresponded to a 3′-truncated form of the gene encoding the first 744 amino acid residues of the protein C-terminally fused to GFP (yielding MsmgΔmmpL3/pMVGH1-mmpL3tb1–744-gfp). Compared to the truncated form of MmpL3 studied by Carel et al.25, MmpL3tb1–744-GFP harbors the last two transmembrane domains of the transporter (TMS11 and TMS12) and is thus only devoid of the cytosolic C-terminal domain of the protein. Consistent with our earlier findings15, the MsmgΔmmpL3 strain rescued with mmpL3tb1–744-gfp was viable indicating that MmpL3tb1–744-GFP was competent at exporting mycolic acids in actively replicating bacilli. Observation of MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp bacilli by fluorescence microscopy revealed a preferential labeling of the poles and septa [Fig. 7, top panels]. Thus, despite expressing the M. tuberculosis ortholog of MmpL3, M. smegmatis was able to successfully recruit the transporter to its poles. Fluorescence in MsmgΔmmpL3/pMVGH1-mmpL3tb1–744-gfp bacilli, in contrast, was dimmer and more diffuse, with reduced accumulation of MmpL3 at the poles and septa, and GFP signals being more evenly distributed along the lateral wall of the bacilli [Fig. 7, lower panels]. In line with Carel et al.’s observations25, the polar localization of MmpL3 was thus reliant on the C-terminal region of the protein, which may now be narrowed down to the last 200 cytoplasmic residues of the transporter.
Loss of polar localization of MmpL3 upon truncation of the C-terminal domain. Representative fluorescence images of MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp (top panels) and MsmgΔmmpL3/pMVGH1-mmpL3tb1–744-gfp (lower panels) showing the focal localization of full-size MmpL3tb-GFP at the old poles and septa of actively dividing bacilli, and dimmer and more diffuse fluorescent signal corresponding to the truncated form of the transporter. The fluorescence intensity of 50 bacilli from each strain (red line) in one representative experiment, plus or minus standard deviation (black lines), was scored and plotted against normalized cell length (right panels).
In an attempt to begin elucidating the molecular mechanisms accounting for the decreased polar localization of MmpL3tb1–744-GFP in M. smegmatis, all the MmpL3 binding partners identified in our two-hybrid system screen were re-tested for interaction with MmpL3tb1–744 using the BACTH assay. The results, which are described on Fig. 3, revealed different patterns of behavior depending on the protein. While some interactions increased in intensity (e.g., Rv0207c, Rv1457c, Rv3064c, Rv3909), Rv0204c, Rv0227c, Rv1337 and Rv3483c appeared, on the contrary, to interact less strongly with MmpL3tb1–744. Changes in interactions with other proteins were not statistically significant. Of note was the decreased interaction between MmpL3tb and MmpL3tb1–744 monomers as well as between MmpL3tb1–744 and MmpL3tb1–744 monomers [Fig. 3] suggesting that the C-terminal end of MmpL3 plays a role in the stable oligomerization of the transporter. This finding may explain why C-terminally truncated forms of MmpL3 were recently reported to crystallize as monomers54,55 rather than as trimers15. Interestingly, no significant differences in binding were detected between CrgA and MmpL3tb full-size, and CrgA and MmpL3tb1–744 [Fig. 3] indicating that the MmpL3/CrgA interaction is not through the C-terminal domain of the transporter. Since the C-terminal domain of MmpL3 is critical to its polar localization, this result suggests that proteins other than CrgA are responsible for the recruitment of the transporter at the old poles and septa. Alternatively or in addition, the decreased interaction between MmpL3 monomers and failure of MmpL3 to properly oligomerize following the truncation of its C-terminal domain may contribute to the more diffuse fluorescence signal in M. smegmatis expressing the truncated MmpL3tb1–744-GFP than those expressing the full-size MmpL3-GFP.
Discussion
Our screen for proteins interacting with MmpL3 identified an unexpectedly low number of hits among enzymes participating in the biosynthesis of mycolic acids. The acetyltransferase TmaT was in fact the only hit in this pathway. Under the conditions of our assays, neither Pks13 nor the mycolyltransferases of the antigen 85 complex that catalyze sequentially close biosynthetic steps upstream and downstream of MmpL3, respectively [Fig. 1], were found to be binding partners. MmpL3, however, was revealed by our screen as a likely protein scaffold for a number of other critically important cell envelope-related biosynthetic machineries including those involved in the assembling and export of peptidoglycan (MviN, CrgA, and possibly Rv3909 and Rv1337), LM, LAM and arabinogalactan (AftD, Rv0227c, Rv1457c), monomeromycolyl diacylglycerol and mycolate wax ester (MmpL11), and perhaps aminoacyl phospholipids (Rv0204c/MprF) [Fig. 8]. Of these, CrgA, Rv0207c, and AftD belonging to various independent functional clusters represented on Fig. 8 were validated by biophysical and/or biochemical methods to physically interact with MmpL3. The existence of a tight network of physically-interacting cell envelope-related pathways was further supported by the detection of interactions between MmpL3 binding partners, e.g., MmpL11/Rv0204c; Rv0227c/TmaT and Rv0227c/AftD [Fig. 4]. Importantly, in addition to interacting indirectly with Wag3120, MmpL3 was also found to interact with the major cell division regulatory protein CrgA pointing to a plausible mechanism through which CrgA, either directly or through its interaction with Wag31 [Fig. 4], may participate in the relocalization of MmpL3 to the septal and polar regions of actively growing cells to promote mycolic acid deposition at these specific sites21,24,25,29. That CrgA is not the only protein contributing to the accumulation of MmpL3 at these specific locations, however, was suggested by the lack of effect of truncating the C-terminal end of the transporter on its ability to interact with CrgA in the BACTH assay, yet, the loss of clear polar localization of the truncated form of MmpL3 in M. smegmatis cells. Collectively, the results presented herein are thus not only suggestive of the existence of a broad connectivity between cell envelope biosynthetic machineries in mycobacteria but also indicate that MmpL3 may serve as an anchor in the coordination of new cell wall deposition with cell elongation and division at the old and septal poles. It is possible that this additional function of MmpL3 contributes to the extreme vulnerability of this therapeutic target56,57.
Network of M. tuberculosis proteins found to interact with MmpL3. Blue lines indicate protein interactions established in the context of the present study. Protein-protein interactions established in the context of previous studies are connected by red lines20,52,53. The functional groups to which the proteins belong or are thought to belong are color-coded. Proteins in yellow circles are related to mycolic acid metabolism. Proteins in the orange circle have not yet been assigned a function. LAM, lipoarabinomannan; LM, lipomannan; AG, arabinogalactan.
Previous studies have reported on the pleiotropic phenotypic effects of silencing or disrupting genes involved in essential mycobacterial cell envelope-related pathways without conclusively determining whether these effects were attributable to compensatory mechanisms, perturbation of the activity of other membrane enzymes and transporters, overall changes in cell envelope permeability, or other mechanisms. For instance the ABC transporter encoded by Rv1458c-Rv1456c was originally proposed to be involved in the translocation of short-chain corynomycolic acids on the basis of the phenotypic characterization of Corynebacterium matruchotii transposon mutants harboring insertions in this transporter58. The transcriptional coupling of this transporter with a mannosyltransferase responsible for the elongation of LM and LAM subsequently led Mishra et al.47 to propose that the altered mycolic acid profile of the C. matruchotii mutants was probably an indirect effect resulting from the loss of mature lipoglycans in the cells. Recently, the high-resolution lipidomics analysis of a C. glutamicum tmaT knock-out mutant revealed an unexpected accumulation of triglycerides in the inner membrane of the mutant that accompanied a decrease in phosphatidylglycerol, alanylated-phosphatidylglycerol and alanylated diacylglycerol59. Likewise, disruption of mmpL11 in M. tuberculosis caused a decrease in cardiolipin content concomitant with alterations in triglyceride levels60,61. Finally, mmpL3 silencing in M. tuberculosis was shown to lead to the up- or down-regulation of 70 genes among which genes encoding a variety of transporters and others involved in mycolic acid and lipid biosynthesis57. The results of our interactome studies strongly suggest that some of the above observations might in fact be the result of perturbations in protein-protein interactions caused by the gene knock-outs simultaneously affecting multiple biosynthetic processes.
It is noteworthy that although several MmpL proteins involved in the export of (glyco)lipids and siderophores in mycobacteria have been found to be genetically and functionally associated with membrane proteins facilitating substrate export (e.g., the ABC-transporter DrrABC, Mycobacteria Membrane Protein Small [MmpS] and small integral membrane proteins of the Gap/Sap family)6,7, no obvious homologs of these proteins are encoded by the genomic region surrounding mmpL3 and no such protein was identified within the MmpL3 interactome reported herein. Our BACTH screen, however, identified a number of integral membrane, lipoproteins and other exported proteins of unknown function (Rv0204c, Rv0227c, Rv0625c, Rv1337, Rv2169c, Rv3064c, Rv3271c, Rv3483, LprC, LppT), some of which may facilitate the export of TMM or other cell envelope constituents. Lipoproteins (other than LprC which did not interact with MmpL3 by SPR), if confirmed to interact with MmpL3, are of particular interest in light of the previous involvement of some of them in the periplasmic translocation of triglycerides, polyketide-derived lipids, glycolipids and lipoglycans62,63,64,65. More studies are required to confirm these potential transporters as binding partners of MmpL3 and to determine the functional significance of these interactions, both from the perspective of mycobacterial physiology and susceptibility to MmpL3 inhibitors.
Methods
Bacterial strains and growth conditions
Escherichia coli DH5α and XL1-Blue, the strains used for cloning purposes, were grown in LB broth and LB agar. M. smegmatis mc2 155 was grown in Middlebrook 7H9-ADC medium supplemented with 0.2% glycerol and 0.05% Tween 80 or on 7H11-ADC agar supplemented with 0.2% Glycerol. For E. coli, kanamycin, ampicillin, spectinomycin and apramycin were used at 50, 100, 50 and 50 μg/mL, respectively. For M. smegmatis, kanamycin, hygromycin and apramycin were used at 25, 50 and 25 μg/mL, respectively.
Two-hybrid system
Bacterial two-hybrid system experiments were conducted using the BACTH (Bacterial Adenylate Cyclase Two-Hybrid) System kit (Euromedex, France). M. tuberculosis mmpL3 and target genes of interest were PCR-amplified and cloned into plasmids pKT25, pKNT25, pUT18 and pUT18C. Genes encoding membrane proteins with known or predicted periplasmic N-terminal ends (aftD, mviN) were also cloned in pUTM18C to redirect the T18 domain to the cytoplasm66. Target genes present in the Mycobacterium tuberculosis Gateway® Clone Set (BEI Resources) were directly subcloned into the Gateway®-compatible plasmids pST25-DEST, pSNT25-DEST or pUT18C-DEST66 using the Gateway® LR Clonase® enzyme (ThermoFisher) as recommended by the manufacturer. Approximately 10 ng of plasmid pairs in different combinations were used to co-transform BTH101 cells and transformants selected on LB agar containing appropriate antibiotics, 0.5 mM IPTG and 40 μg/mL X-Gal. Plates were incubated at 30 °C for 48 h prior to visual inspection for blue colonies indicative of protein interactions.
For the screening of a genomic library of M. tuberculosis open reading frames against mmpL3, we used the M. tuberculosis Gateway® Clone Set from BEI Resources (NR-19274) consisting of 3,724 unique M. tuberculosis H37Rv and CDC1551 ORFs cloned in vector pDONR™221 in E. coli DH10B-T1 cells. The forty-two 96-well plates carrying these clones were re-grown in LB-kanamycin medium after which the 96-wells from each plate were combined and the plasmids extracted, thereby yielding 42 pools of 96 plasmids. The M. tuberculosis ORFs from each plasmid pool were then subcloned into pST25-DEST and pSNT25-DEST using the Gateway® LR Clonase® enzyme as described above. LR Clonase reactions were next used to transform BTH101 competent cells harboring either pUT18::mmpL3 or pUT18C::mmpL3, and the transformation mixtures plated on five to ten LB plates containing ampicillin, spectinomycin, IPTG and X-Gal in order to select for blue colonies indicative of protein interactions. Plasmids purified from blue colonies were confirmed for positive interaction by co-transformation with empty pUT18 or pUT18C plasmids (negative controls) or plasmids harboring mmpL3-T18 fusions (positive controls), and finally sequenced to identify the genes yielding positive interactions with mmpL3.
All pairwise co-transformants developing a blue color in the BATCH screen were grown in LB broth, and the cultures processed for β-galactosidase activity as described previously67. Briefly, 3 independent colonies per pairwise co-transformation were picked and grown overnight in 96-well plates containing LB medium supplemented with antibiotics and 0.5 mM IPTG. 50 μl of each culture were used to determine OD600nm while another 200 μl was transferred into 96-well propylene blocks containing 800 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4 and 50 mM β-mercaptoethanol). After adding 20 μl of freshly prepared 0.1% SDS and 40 μl of chloroform to permeabilize the cells, 50 μl aliquots of the upper aqueous layer were transferred into 96-well flat-bottom microplates containing 150 μl of Z buffer and pre-equilibrated at 28 °C in the microplate reader. 40 μl of 0.4% ONPG were finally dispensed and the β-galactosidase reaction carried out at 28 °C for 40 min with measurement of the OD420nm every 2 min. The relative β-galactosidase activity is calculated by the formula [(OD420nm at time T2 − OD420nm at time T1)/(T2 − T1)]/OD600nm. The T2 and T1 time points are chosen to be in the linear part of the kinetic.
Construction of M. smegmatis strains expressing full-size and C-terminal truncated forms of MmpL3 fused to GFP
MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp and MsmgΔmmpL3/pMVGH1-mmpL3tb1–744-gfp were generated as described previously15. Briefly, pMVGH14 constructs expressing the mmpL3 gene from M. tuberculosis, either full-size or truncated at its 3′-end so as to only express the first 744 amino acid residues of the protein, and fused to gfp and a C-terminal hexahistidine tag were used to transform a M. smegmatis strain having undergone a single crossover event at its mmpL3 locus4. MsmgΔmmpL3 allelic exchange mutants expressing either mmpL3tb1–744-gfp or mmpL3tb-gfp were then selected by plating on 7H11-ADC agar containing Kan, Hyg and sucrose as described15.
Fluorescence microscopy
MsmgΔmmpL3/pMVGH1-mmpL3tb-gfp and MsmgΔmmpL3/pMVGH1-mmpL3tb1–744-gfp cultures grown to exponential phase were collected, washed twice in phosphate-buffered saline containing 0.05% Tween 80, and fixed in freshly prepared 2% paraformaldehyde for 30 min at room temperature. Approximately 106 cells were next transferred to a glass slide by Cytospin, mounted with Fluoro-Gel (Electron Microscopy Science) and visualized using a KEYENCE BZ-X700 fluorescence microscope. Fluorescent images for the two strains were acquired at identical exposure times. Multiple independent experiments were performed and data from one representative experiment were used for statistical analysis.
Production and purification of CrgA, Rv0207c, LprC, Pks13, MviN and AftD
Recombinant forms of the CrgA, Rv0207c and Pks13 proteins from M. tuberculosis H37Rv harboring C-terminal hexahistidine tags were produced in E. coli Rosetta (DE3) using the pET29a (CrgA, Rv0207c) or pET26b (Pks13) expression systems (EMD Biosciences). AftD from Mycobacterium abscessus (1410 amino acids; 63% identity, 74% similarity to AftD from M. tuberculosis on a 1397 amino acid overlap) was expressed in E. coli BL21 (DE3) pLysS cells using a pNYCOMPS-N23 plasmid with a N-terminal 10-histidine tag. A recombinant form of LprC devoid of its first 21 amino acids (so as to remove the signal peptide of the protein including residue Cys21 predicted to be acylated) and harboring an N-terminal hexahistidine tag was produced in E. coli CR43 (DE3) using the pET14b expression system (EMD Biosciences). Similarly, E. coli C43 (DE3) cells harboring pET29a::MviN were utilized for purification of the His-tagged MviN. Overnight grown cells were sub-cultured 1:100 in fresh LB plus drug, and grown to OD600 0.6–0.8, and then induced with 0.1 mM IPTG for four hours at 37 °C (CrgA, Pks13 and MviN), overnight at 16 °C (LprC and Rv0207c) or overnight at 22 °C (AftD). Cells were harvested by centrifugation at 4,000 × g for 20 min, then resuspended in lysis buffer (50 mM Tris pH 8.0, 5 mM EDTA, 1 mM PMSF) and broken by sonication. For AftD, the lysis buffer used was 20 mM HEPES pH 7.5, 200 mM NaCl, 20 mM MgSO4, 10 μg/mL DNase I (Roche), 8 μg/mL RNase A (Roche), 1 mM tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 1 mM PMSF, 1 tablet/1.5 L buffer EDTA-free complete protease inhibitor cocktail (Roche). Unbroken cells were removed by centrifugation at 4,000 × g, and then membranes were collected by ultracentrifugation at 40,000 × g at 4 °C for 1 hour. Membranes were solubilized with HS buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 2% Triton X-100) overnight at 4 °C with stirring. Insoluble material was removed by ultracentrifugation at 40,000 × g at 4 °C for 1 hour. Soluble samples were passed through a SP-HP column cation-exchange column (GE Healthcare) equilibrated in HS buffer, and flow-through was adjusted to 20 mM imidazole and 200 mM NaCl final concentration before loading onto HIS-bind column. The column was washed extensively with HW buffer (20 mM HEPES pH 7.4, 150 mM NaCl, 0.2% Triton X-100 supplemented with 20 mM and 50 mM imidazole; 60 mM imidazole for AftD), before elution (HW supplemented with 500 mM imidazole; 300 mM imidazole for AftD). Elution fractions were dialyzed against SPR Assay Buffer (20 mM HEPES pH 7.4, 150 mM NaCl, and 0.2% Triton X-100) and concentrated by ultrafiltration before SPR as necessary. 0.03% n-Dodecyl-β-D-maltoside (DDM), instead of 0.2% Triton X-100, was used in the SPR assay buffer in the case of AftD.
Purification of MmpL3 and SPR experiments
MsmgΔmmpL3/pMVGH1-mmpL3tb cells (expressing a C-terminally hexahistidine-tagged form of mmpL3 from M. tuberculosis devoid of gfp fusion) were grown in four liters of 7H9-ADC medium supplemented with 0.2% glycerol, 0.05% Tween-80, 25 μg/mL kanamycin and 50 μg/mL hygromycin B for 48 hours at 30 °C with shaking. The MmpL3 protein was purified and SPR experiments were carried out as described previously68. The protein analytes in SPR assay buffer were injected over immobilized MmpL3 at 10 μL/min for 30 sec and then allowed to disassociate for 180 sec. Data was collected at 10 Hz and 25 °C. Similar results were obtained using Bio-layer interferometry (data not shown).
Co-immunoprecipitation
crgA from M. tuberculosis H37Rv was cloned into pFAX, a mycobacterial integrative plasmid harboring an apramycin resistance cassette engineered in-house to allow for the expression of recombinant proteins N-terminally-fused to the FLAG epitope from the hsp60 promoter. M. smegmatis ΔmmpL3 cells co-expressing mmpL3tb-gfp from pMVGH1-mmpL3tb-gfp and crgA-FLAG from pFAX-crgA grown to an OD of ~0.8 were treated with 1.25 mM of the cross-linking agent dithiobis[succinimidyl propionate] (DSP; Thermo Scientific) at 37 °C for 30 min in PBS. Upon incubation with the cross-linker, bacteria were harvested, lysed by bead-beating in lysis buffer consisting of 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10 μg/ml DNAse I and proteinase inhibitor cocktail (Sigma-Aldrich), and the membranes collected by ultracentrifugation and resuspended in a buffer containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl, 10% glycerol and 1% DDM (GoldBio). Upon solubilization of the membrane pellets overnight on ice, the insoluble material was removed by centrifugation at 27,000 × g at 4 °C for 30 min, and solubilized MmpL3tb and its binding partners purified on Ni-NTA agarose resin (ThermoFisher). Upon copious washing of the resin with washing buffer (50 mM Tris-HCl pH 8.0, 400 mM NaCl, 5% glycerol, 20 mM imidazole) to remove unbound proteins, cross-linked protein complexes were eluted from the column with elution buffer (50 mM Tris-HCl pH 8.0, 400 mM NaCl, 5% glycerol, 250 mM imidazole, 0.1% DDM). Elution fractions were run in 4–12% NuPAGE Bis-Tris Gels and the gels analyzed by in-gel fluorescence (to detect MmpL3tb-GFP) followed by Coomassie blue staining. DSP-induced complexes were reversed by incubation with dithiothreitol prior to loading on SDS-PAGE. Immunoblot analysis used anti-FLAG M2 antibodies (Sigma-Aldrich) as primary antibodies, and an anti-mouse IgG antibody coupled to horseradish peroxidase (HRP) (Sigma-Aldrich) as the secondary antibody. Radiance HRP substrate (Azure Biosystems) followed by chemiluminescence detection was used to detect the FLAG-tagged CrgA protein.
References
North, E. J., Jackson, M. & Lee, R. E. New approaches to target the mycolic acid biosynthesis pathway for the development of tuberculosis therapeutics. Curr Pharm Des 20, 4357–4378 (2014).
Li, W. et al. MmpL3 as a Target for the Treatment of Drug-Resistant Nontuberculous Mycobacterial Infections. Front Microbiol 9, 1547 (2018).
Tahlan, K. et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob Agents Chemother 56, 1797–1809 (2012).
Grzegorzewicz, A. E. et al. Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat Chem Biol 8, 334–341 (2012).
Xu, Z., Meshcheryakov, V. A., Poce, G. & Chng, S. S. MmpL3 is the flippase for mycolic acids in mycobacteria. Proc Natl Acad Sci USA 114, 7993–7998 (2017).
Chalut, C. MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis (Edinb) 100, 32–45 (2016).
Touchette, M. H. & Seeliger, J. C. Transport of outer membrane lipids in mycobacteria. Biochim Biophys Acta 1862, 1340–1354 (2017).
Zheng, J. et al. Combining blue native polyacrylamide gel electrophoresis with liquid chromatography tandem mass spectrometry as an effective strategy for analyzing potential membrane protein complexes of Mycobacterium bovis bacillus Calmette-Guerin. BMC genomics 12, 40 (2011).
Veyron-Churlet, R., Guerrini, O., Mourey, L., Daffé, M. & Zerbib, D. Protein-protein interactions within the fatty acid synthase-II system of Mycobacterium tuberculosis are essential for mycobacterial viability. Mol Microbiol 54, 1161–1172 (2004).
Veyron-Churlet, R. et al. The biosynthesis of mycolic acids in Mycobacterium tuberculosis relies on multiple specialized elongation complexes interconnected by specific protein-protein interactions. J Mol Biol 353, 847–858 (2005).
Cantaloube, S., Veyron-Churlet, R., Haddache, N., Daffé, M. & Zerbib, D. The Mycobacterium tuberculosis FAS-II dehydratases and methyltransferases define the specificity of the mycolic acid elongation complexes. PLoS ONE 6, e29564 (2011).
Lefebvre, C. et al. HadD, a novel fatty acid synthase type II protein, is essential for alpha- and epoxy-mycolic acid biosynthesis and mycobacterial fitness. Sci Rep 8, 6034 (2018).
Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M. & Engelhardt, H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 105, 3963–3967 (2008).
Zuber, B. et al. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 190, 5672–5680 (2008).
Belardinelli, J. M. et al. Structure-Function Profile of MmpL3, the Essential Mycolic Acid Transporter from Mycobacterium tuberculosis. ACS Infect Dis 2, 702–713 (2016).
Fay, A. et al. Two accessory proteins govern mycolic acid transport in mycobacteria. bioRxiv, https://doi.org/10.1101/581447 (2019).
Nikaido, H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzymol Relat Areas Mol Biol 77, 1–60 (2011).
Thanky, N. R., Young, D. B. & Robertson, B. D. Unusual features of the cell cycle in mycobacteria: polar-restricted growth and the snapping-model of cell division. Tuberculosis (Edinb) 87, 231–236 (2007).
Aldridge, B. B. et al. Asymmetry and aging of mycobacterial cells lead to variable growth and antibiotic susceptibility. Science 335, 100–104 (2012).
Xu, W. X. et al. The Wag31 protein interacts with AccA3 and coordinates cell wall lipid permeability and lipophilic drug resistance in Mycobacterium smegmatis. Biochem Biophys Res Commun 448, 255–260 (2014).
Meniche, X. et al. Subpolar addition of new cell wall is directed by DivIVA in mycobacteria. Proc Natl Acad Sci USA 111, E3243–3251 (2014).
Hayashi, J. M. et al. Spatially distinct and metabolically active membrane domain in mycobacteria. Proc Natl Acad Sci USA 113, 5400–5405 (2016).
Hayashi, J. M. et al. Stress-Induced Reorganization of the Mycobacterial Membrane Domain. MBio 9 (2018).
Puffal, J., Garcia-Heredia, A., Rahlwes, K. C., Siegrist, M. S. & Morita, Y. S. Spatial control of cell envelope biosynthesis in mycobacteria. Pathog Dis 76 (2018)
Carel, C. et al. Mycobacterium tuberculosis proteins involved in mycolic acid synthesis and transport localize dynamically to the old growing pole and septum. PLoS ONE 9, e97148 (2014).
Backus, K. M. et al. Uptake of unnatural trehalose analogs as a reporter for Mycobacterium tuberculosis. Nat Chem Biol 7, 228–235 (2011).
Swarts, B. M. et al. Probing the mycobacterial trehalome with bioorthogonal chemistry. J Am Chem Soc 134, 16123–16126 (2012).
Foley, H. N., Stewart, J. A., Kavunja, H. W., Rundell, S. R. & Swarts, B. M. Bioorthogonal Chemical Reporters for Selective InSitu Probing of Mycomembrane Components in Mycobacteria. Angew Chem Int Edit 55, 2053–2057 (2016).
Hodges, H. L., Brown, R. A., Crooks, J. A., Weibel, D. B. & Kiessling, L. L. Imaging mycobacterial growth and division with a fluorogenic probe. Proc Natl Acad Sci USA 115, 5271–5276 (2018).
Karimova, G., Pidoux, J., Ullmann, A. & Ladant, D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA 95, 5752–5756 (1998).
Baulard, A. R. et al. In vivo interaction between the polyprenol phosphate mannose synthase Ppm1 and the integral membrane protein Ppm2 from Mycobacterium smegmatis revealed by a bacterial two-hybrid system. J Biol Chem 278, 2242–2248 (2003).
Clarke, B. R., Greenfield, L. K., Bouwman, C. & Whitfield, C. Coordination of polymerization, chain termination, and export in assembly of the Escherichia coli lipopolysaccharide O9a antigen in an ABC-transporter-dependent pathway. J Biol Chem 284, 30662–30672 (2009).
Yamaryo-Botte, Y. et al. Acetylation of trehalose mycolates is required for efficient MmpL-mediated membrane transport in Corynebacterineae. ACS Chem Biol 10, 734–746 (2015).
Slavetinsky, C., Kuhn, S. & Peschel, A. Bacterial aminoacyl phospholipids - Biosynthesis and role in basic cellular processes and pathogenicity. Biochim Biophys Acta 1862, 1310–1318 (2017).
Pacheco, S. A., Hsu, F. F., Powers, K. M. & Purdy, G. E. MmpL11 protein transports mycolic acid-containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. J Biol Chem 288, 24213–24222 (2013).
Tullius, M. V. et al. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc Natl Acad Sci USA 108, 5051–5056 (2011).
Owens, C. P. et al. The Mycobacterium tuberculosis secreted protein Rv0203 transfers heme to membrane proteins MmpL3 and MmpL11. J Biol Chem 288, 21714–21728 (2013).
Camacho, L. R., Ensergueix, D., Pérez, E., Gicquel, B. & Guilhot, C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol 34, 257–267 (1999).
Sassetti, C. M. & Rubin, E. J. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. USA 100, 12989–12994 (2003).
Cashmore, T. J. et al. Identification of a Membrane Protein Required for Lipomannan Maturation and Lipoarabinomannan Synthesis in Corynebacterineae. J Biol Chem 292, 4976–4986 (2017).
Škovierová, H. et al. AftD, a novel essential arabinofuranosyltransferase from mycobacteria. Glycobiology 19, 1235–1247 (2009).
Alderwick, L. J. et al. AftD functions as an alpha1–>5 arabinofuranosyltransferase involved in the biosynthesis of the mycobacterial cell wall core. Cell Surf 1, 2–14 (2018).
Gee, C. L. et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal 5, ra7 (2012).
Song, H., Sandie, R., Wang, Y., Andrade-Navarro, M. A. & Niederweis, M. Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis 88, 526–544 (2008).
Kateete, D. P. et al. Rhomboid homologs in mycobacteria: insights from phylogeny and genomic analysis. BMC Microbiol 10, 272 (2010).
Kateete, D. P. et al. Rhomboids of Mycobacteria: characterization using an aarA mutant of Providencia stuartii and gene deletion in Mycobacterium smegmatis. PLoS ONE 7, e45741 (2012).
Mishra, A. K. et al. Identification of a novel alpha(1->6) mannopyranosyltransferase MptB from Corynebacterium glutamicum by deletion of a conserved gene, NCgl1505, affords a lipomannan- and lipoarabinomannan-deficient mutant. Mol Microbiol 68, 1595–1613 (2008).
Wu, K. J. et al. Characterization of Conserved and Novel Septal Factors in Mycobacterium smegmatis. J Bacteriol 200 (2018).
Daffé, M., Crick, D. C. & Jackson, M. Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol Spectrum 2, MGM2-0021–2013 (2014).
Grzegorzewicz, A. E. et al. Assembling of the Mycobacterium tuberculosis Cell Wall Core. J Biol Chem 291, 18867–18879 (2016).
Xu, J. et al. A unique Mycobacterium ESX-1 protein co-secretes with CFP-10/ESAT-6 and is necessary for inhibiting phagosome maturation. Mol Microbiol 66, 787–800 (2007).
Plocinski, P. et al. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J Bacteriol 193, 3246–3256 (2011).
Plocinski, P. et al. Mycobacterium tuberculosis CwsA interacts with CrgA and Wag31, and the CrgA-CwsA complex is involved in peptidoglycan synthesis and cell shape determination. J Bacteriol 194, 6398–6409 (2012).
Zhang, B. et al. Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target. Cell 176, 636–648 e613 (2019).
Su, C. C. et al. MmpL3 is a lipid transporter that binds trehalose monomycolate and phosphatidylethanolamine. Proc Natl Acad Sci USA, https://doi.org/10.1073/pnas.1901346116 (2019).
Li, W. et al. Therapeutic Potential of the Mycobacterium tuberculosis Mycolic Acid Transporter, MmpL3. Antimicrob Agents Chemother 60, 5198–5207 (2016).
Degiacomi, G. et al. Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci Rep 7, 43495 (2017).
Wang, C., Hayes, B., Vestling, M. M. & Takayama, K. Transposome mutagenesis of an integral membrane transporter in Corynebacterium matruchotii. Biochem Biophys Res Commun 340, 953–960 (2006).
Klatt, S. et al. Identification of novel lipid modifications and intermembrane dynamics in Corynebacterium glutamicum using high-resolution mass spectrometry. J Lipid Res 59, 1190–1204 (2018).
Wright, C. C. et al. The Mycobacterium tuberculosis MmpL11 Cell Wall Lipid Transporter Is Important for Biofilm Formation, Intracellular Growth, and Nonreplicating Persistence. Infect Immun 85 (2017).
Bothra, A. et al. Phospholipid homeostasis, membrane tenacity and survival of Mtb in lipid rich conditions is determined by MmpL11 function. Sci Rep 8, 8317 (2018).
Sulzenbacher, G. et al. LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J 25, 1436–1444 (2006).
Shukla, S. et al. Mycobacterium tuberculosis lipoprotein LprG binds lipoarabinomannan and determines its cell envelope localization to control phagolysosomal fusion. PLoS Pathog 10, e1004471 (2014).
Gaur, R. L. et al. LprG-mediated surface expression of lipoarabinomannan is essential for virulence of Mycobacterium tuberculosis. PLoS Pathog 10, e1004376 (2014).
Martinot, A. J. et al. Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis. PLoS Pathog 12, e1005351 (2016).
Ouellette, S. P., Gauliard, E., Antosova, Z. & Ladant, D. A Gateway((R)) -compatible bacterial adenylate cyclase-based two-hybrid system. Environ Microbiol Rep 6, 259–267 (2014).
Battesti, A. & Bouveret, E. The bacterial two-hybrid system based on adenylate cyclase reconstitution in Escherichia coli. Methods 58, 325–334 (2012).
Li, W. et al. Direct Inhibition of MmpL3 by Novel Antitubercular Compounds. ACS Infect Dis, https://doi.org/10.1021/acsinfecdis.9b00048 (2019).
Marrakchi, H., Lanéelle, M. A. & Daffé, M. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol 21, 67–85 (2014).
Acknowledgements
This work was supported by a grant from the National Institutes of Health/National Institute of Allergy and Infectious Diseases grants AI116525 (to M.J and H.I.Z). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Dr. Daniel Ladant (Institut Pasteur, Paris, France) for the BACTH expression plasmids, Dr. A. Quėmard (IPBS-CNRS, Toulouse, France) for the pks13 expression plasmid, and Dr. Heather Hodges for her help with fluorescence imaging. We thank the Colorado State University Libraries Open Access Research and Scholarship Fund for their partial support of publication fees. The following reagents were obtained through BEI Resources, NIAID, NIH: (i) Mycobacterium tuberculosis Gateway® Clone Set, Recombinant in Escherichia coli, Plates 1–42, (NR-19274); and (ii) Ag85 Complex, Purified Native Proteins from Mycobacterium tuberculosis, Strain H37Rv (NR-14855). SPR experiments were carried out using a Biacore T200 instrument of the Oklahoma Medical Research Foundation Biacore Facility, which is funded by the Shared Instrumentation Grant S10 OD025014.
Author information
Authors and Affiliations
Contributions
J.M.B., W.L., H.I.Z. and M.J. designed research. J.M.B., W.L., C.M.S., Y.Z.T. and V.J. performed research. J.M.B., W.L., C.M.S., H.I.Z. and M.J. analyzed data. H.I.Z., J.M.B., F.M. and M.J. wrote the main manuscript text. J.M.B., W.L. and C.M.S. prepared the figures. All authors reviewed the final version of the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Belardinelli, J.M., Stevens, C.M., Li, W. et al. The MmpL3 interactome reveals a complex crosstalk between cell envelope biosynthesis and cell elongation and division in mycobacteria. Sci Rep 9, 10728 (2019). https://doi.org/10.1038/s41598-019-47159-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47159-8
This article is cited by
-
In vivo imaging of MmpL transporters reveals distinct subcellular locations for export of mycolic acids and non-essential trehalose polyphleates in the mycobacterial outer membrane
Scientific Reports (2023)
-
Novel chemical entities inhibiting Mycobacterium tuberculosis growth identified by phenotypic high-throughput screening
Scientific Reports (2022)
-
The role of transport proteins in the production of microbial glycolipid biosurfactants
Applied Microbiology and Biotechnology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.