Spatial Referencing of Hyperspectral Images for Tracing of Plant Disease Symptoms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material & Fungal Pathogens
2.1.1. Plant Material
2.1.2. Fungal Pathogens
2.2. Hyperspectral Measurements
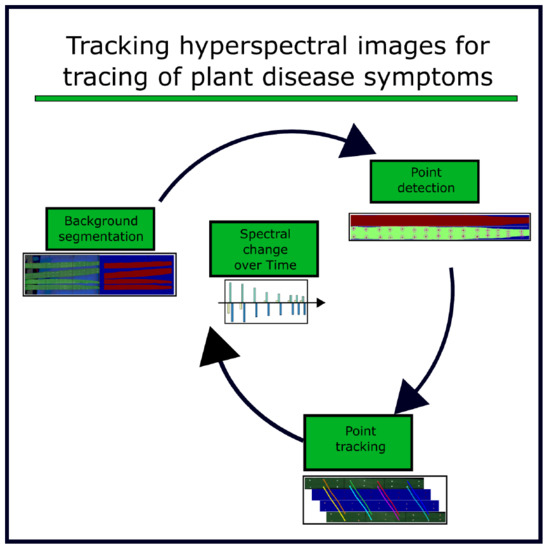
2.3. Algorithm for Hyperspectral Image Referencing
2.3.1. Background Segmentation
2.3.2. Reference Point Extraction
2.3.3. Assignment of Reference Points
2.3.4. Spatial Image Transformation Models
2.3.5. Evaluation of Transformation Accuracy
2.4. Vegetation Indices
2.5. Presymptomatic Labeling
3. Results
3.1. Background Segmentation and Reference Point Detection
3.2. Transformation Model
3.3. Presymptomatic Labeling
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ARI | Anthocyanin Reflectance Index |
| LWM | Local Weighted Mean |
| NDVI | Normalized Difference Vegetation Index |
| PRI | Photochemical Reflectance Index |
| RMSE | Root Mean Square Error |
| UAV | Unmanned Aerial Vehicle |
| VISNIR | Visual-nearinfrared |
References
- Mahlein, A.K.; Kuska, M.; Behmann, J.; Polder, G.; Walter, A. Hyperspectral sensors and imaging technologies in phytopathology: State of the art. Annu. Rev. Phytopathol. 2018, 56, 535–558. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, G.A. Hyperspectral remote sensing of plant pigments. J. Exp. Bot. 2006, 58, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bock, C.; Poole, G.; Parker, P.; Gottwald, T. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Mahlein, A.K. Plant disease detection by imaging sensors–parallels and specific demands for precision agriculture and plant phenotyping. Plant Dis. 2016, 100, 241–251. [Google Scholar] [CrossRef]
- Kuska, M.; Wahabzada, M.; Leucker, M.; Dehne, H.W.; Kersting, K.; Oerke, E.C.; Steiner, U.; Mahlein, A.K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28. [Google Scholar] [CrossRef] [PubMed]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Aasen, H.; Burkart, A.; Bolten, A.; Bareth, G. Generating 3D hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. ISPRS J. Photogramm. Remote Sens. 2015, 108, 245–259. [Google Scholar] [CrossRef]
- Honkavaara, E.; Rosnell, T.; Oliveira, R.; Tommaselli, A. Band registration of tuneable frame format hyperspectral UAV imagers in complex scenes. ISPRS J. Photogramm. Remote Sens. 2017, 134, 96–109. [Google Scholar] [CrossRef]
- Burkart, A.; Aasen, H.; Alonso, L.; Menz, G.; Bareth, G.; Rascher, U. Angular dependency of hyperspectral measurements over wheat characterized by a novel UAV based goniometer. Remote Sens. 2015, 7, 725–746. [Google Scholar] [CrossRef] [Green Version]
- Rouse, J., Jr.; Haas, R.; Schell, J.; Deering, D. Monitoring vegetation systems in the Great Plains with ERTS. In Proceedings of the Third Earth Resources Technology Satellite-1 Symposium, Washington, DC, USA, 10–14 December 1973. [Google Scholar]
- Bockus, W.W.; Bowden, R.; Hunger, R.; Murray, T.; Smiley, R. Compendium of Wheat Diseases and Pests, 3rd ed.; American Phytopathological Society (APS Press): Sao Paulo, MN, USA, 2010. [Google Scholar]
- Camargo, A.; Smith, J. An image-processing based algorithm to automatically identify plant disease visual symptoms. Biosyst. Eng. 2009, 102, 9–21. [Google Scholar] [CrossRef]
- West, J.S.; Bravo, C.; Oberti, R.; Lemaire, D.; Moshou, D.; McCartney, H.A. The potential of optical canopy measurement for targeted control of field crop diseases. Annu. Rev. Phytopathol. 2003, 41, 593–614. [Google Scholar] [CrossRef] [PubMed]
- Bravo, C.; Moshou, D.; West, J.; McCartney, A.; Ramon, H. Early Disease Detection in Wheat Fields using Spectral Reflectance. Biosyst. Eng. 2003, 84, 137–145. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Rumpf, T.; Römer, C.; Plümer, L. A review of advanced machine learning methods for the detection of biotic stress in precision crop protection. Precis. Agric. 2015, 16, 239–260. [Google Scholar] [CrossRef]
- Hartley, R.; Zisserman, A. Multiple View Geometry in Computer Vision; Cambridge University Press: Cambridge, UK, 2003. [Google Scholar]
- Zitova, B.; Flusser, J. Image registration methods: A survey. Image Vis. Ccomput. 2003, 21, 977–1000. [Google Scholar] [CrossRef]
- Salvi, J.; Matabosch, C.; Fofi, D.; Forest, J. A review of recent range image registration methods with accuracy evaluation. Image Vis. Comput. 2007, 25, 578–596. [Google Scholar] [CrossRef]
- Eling, C.; Klingbeil, L.; Kuhlmann, H. Real-time single-frequency GPS/MEMS-IMU attitude determination of lightweight UAVs. Sensors 2015, 15, 26212–26235. [Google Scholar] [CrossRef] [PubMed]
- Toutin, T. Geometric processing of remote sensing images: Models, algorithms and methods. Int. J. Remote Sens. 2004, 25, 1893–1924. [Google Scholar] [CrossRef]
- Gwo, C.Y.; Wei, C.H. Plant identification through images: Using feature extraction of key points on leaf contours1. Appl. Plant Sci. 2013, 1, 1200005. [Google Scholar] [CrossRef]
- Mouine, S.; Yahiaoui, I.; Verroust-Blondet, A. Combining leaf salient points and leaf contour descriptions for plant species recognition. In Proceedings of the International Conference Image Analysis and Recognition, Povoa do Varzim, Portugal, 26–28 June 2013; Springer: Berlin, Germany, 2013; pp. 205–214. [Google Scholar]
- Kolivand, H.; Fern, B.M.; Rahim, M.S.M.; Sulong, G.; Baker, T.; Tully, D. An expert botanical feature extraction technique based on phenetic features for identifying plant species. PLoS ONE 2018, 13, e0191447. [Google Scholar] [CrossRef]
- Gupta, M.D.; Nath, U. Divergence in patterns of leaf growth polarity is associated with the expression divergence of miR396. Plant Cell 2015. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Paulus, S.; Kuhlmann, H.; Oerke, E.C.; Plümer, L. Calibration of hyperspectral close-range pushbroom cameras for plant phenotyping. ISPRS J. Photogramm. Remote Sens. 2015, 106, 172–182. [Google Scholar] [CrossRef]
- Behmann, J.; Mahlein, A.K.; Paulus, S.; Dupuis, J.; Kuhlmann, H.; Oerke, E.C.; Plümer, L. Generation and application of hyperspectral 3D plant models: Methods and challenges. Mach. Vis. Appl. 2016, 27, 611–624. [Google Scholar] [CrossRef]
- De Vylder, J.; Douterloigne, K.; Vandenbussche, F.; Van Der Straeten, D.; Philips, W. A non-rigid registration method for multispectral imaging of plants. In Proceedings of the 2012 SPIE Defense, Security, and Sensing, Baltimore, MD, USA, 23–27 April 2012; Volume 8369, p. 836907. [Google Scholar]
- Bar-Sinai, Y.; Julien, J.D.; Sharon, E.; Armon, S.; Nakayama, N.; Adda-Bedia, M.; Boudaoud, A. Mechanical stress induces remodeling of vascular networks in growing leaves. PLoS Comput. Boil. 2016, 12, e1004819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balduzzi, M.; Binder, B.M.; Bucksch, A.; Chang, C.; Hong, L.; Iyer-Pascuzzi, A.S.; Pradal, C.; Sparks, E.E. Reshaping plant biology: Qualitative and quantitative descriptors for plant morphology. Front. Plant Sci. 2017, 8, 117. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yang, W.; Wheaton, A.; Cooley, N.; Moran, B. Efficient registration of optical and IR images for automatic plant water stress assessment. Comput. Electron. Agric. 2010, 74, 230–237. [Google Scholar] [CrossRef]
- Henke, M.; Junker, A.; Neumann, K.; Altmann, T.; Gladilin, E. Automated alignment of multi-modal plant images using integrative phase correlation approach. Front. Plant Sci. 2018, 9, 1519. [Google Scholar] [CrossRef] [PubMed]
- Lowe, D.G. Distinctive image features from scale-invariant keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Yin, X.; Liu, X.; Chen, J.; Kramer, D.M. Multi-leaf alignment from fluorescence plant images. In Proceedings of the IEEE 2014 IEEE Winter Conference on Applications of Computer Vision (WACV), Steamboat Springs, CO, USA, 24–26 March 2014; pp. 437–444. [Google Scholar]
- Raza, S.E.A.; Sanchez, V.; Prince, G.; Clarkson, J.P.; Rajpoot, N.M. Registration of thermal and visible light images of diseased plants using silhouette extraction in the wavelet domain. Pattern Recognit. 2015, 48, 2119–2128. [Google Scholar] [CrossRef]
- Raza, S.E.A.; Prince, G.; Clarkson, J.P.; Rajpoot, N.M. Automatic detection of diseased tomato plants using thermal and stereo visible light images. PLoS ONE 2015, 10, e0123262. [Google Scholar] [CrossRef]
- Luhmann, T.; Robson, S.; Kyle, S.; Harley, I. Close Range Photogrammetry: Principles, Techniques and Applications; Whittles: Dunbeath, UK, 2006. [Google Scholar]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Meier, U. Growth Stages of Mono-and Dicotyledonous Plants; Blackwell, Wissenschafts-Verlag: Berlin, Germany, 1997. [Google Scholar]
- Grahn, H.; Geladi, P. Techniques and Applications of Hyperspectral Image Analysis; John Wiley Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Goshtasby, A. Image registration by local approximation methods. Image Vis. Comput. 1988, 6, 255–261. [Google Scholar] [CrossRef]
- Gamon, J.; Penuelas, J.; Field, C. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical properties and nondestructive estimation of anthocyanin content in plant leaves. Photochem. Photobiol. 2001, 74, 38. [Google Scholar] [CrossRef]
- Wang, E.; Dong, C.; Park, R.F.; Roberts, T.H. Carotenoid pigments in rust fungi: Extraction, separation, quantification and characterisation. Fungal Boil. Rev. 2018, 32, 166–180. [Google Scholar] [CrossRef]
- Rumpf, T.; Mahlein, A.K.; Steiner, U.; Oerke, E.C.; Dehne, H.W.; Plümer, L. Early detection and classification of plant diseases with Support Vector Machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Kuska, M.T.; Behmann, J.; Grosskinsky, D.K.; Roitsch, T.; Mahlein, A.K. Screening of barley resistance against powdery mildew by simultaneous high-throughput enzyme activity signature profiling and multispectral imaging. Front. Plant Sci. 2018, 9, 1074. [Google Scholar] [CrossRef]
- Lazebnik, S.; Schmid, C.; Ponce, J. A sparse texture representation using local affine regions. IEEE Trans. Pattern Anal. Mach. Intell. 2005, 27, 1265–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prusinkiewicz, P.; Lindenmayer, A. The Algorithmic Beauty of Plants; Springer: Berlin/Heidelberg, Germany, 1996. [Google Scholar]
- Vos, J.; Evers, J.B.; Buck-Sorlin, G.H.; Andrieu, B.; Chelle, M.; de Visser, P.H.B. Functional-structural plant modelling: A new versatile tool in crop science. J. Exp. Bot. 2009, 61, 2101–2115. [Google Scholar] [CrossRef] [Green Version]






| Similarity | Affine | Projective | Polynomial | LWM | |
|---|---|---|---|---|---|
| Accuracy (px) | 1.24 (0.75) | 1.23 (0.76) | 1.09 (0.75) | 0.26 (0.092) | 0.19 (0.07) |
| Stability (px) | 1.06 (0.62) | 1.08 (0.61) | 0.97 (0.60) | 0.47 (0.17) | 0.36 (0.15) |
| Extrapolation (px) | 2.61 (1.57) | 2.61 (1.59) | 2.37 (1.59) | 0.83 (0.31) | 0.76 (0.30) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Behmann, J.; Bohnenkamp, D.; Paulus, S.; Mahlein, A.-K. Spatial Referencing of Hyperspectral Images for Tracing of Plant Disease Symptoms. J. Imaging 2018, 4, 143. https://doi.org/10.3390/jimaging4120143
Behmann J, Bohnenkamp D, Paulus S, Mahlein A-K. Spatial Referencing of Hyperspectral Images for Tracing of Plant Disease Symptoms. Journal of Imaging. 2018; 4(12):143. https://doi.org/10.3390/jimaging4120143
Chicago/Turabian StyleBehmann, Jan, David Bohnenkamp, Stefan Paulus, and Anne-Katrin Mahlein. 2018. "Spatial Referencing of Hyperspectral Images for Tracing of Plant Disease Symptoms" Journal of Imaging 4, no. 12: 143. https://doi.org/10.3390/jimaging4120143