The Pedological Context Modulates the Response of Soil Microbial Communities to Agroecological Management
- 1Ecology of Plants, Institute of Biology, Freie Universität Berlin, Berlin, Germany
- 2AgricultureIsLife, TERRA Teaching and Research Centre, Gembloux Agro-Bio Tech, University of Liège, Liège, Belgium
- 3Biodiversity and Landscapes Unit, TERRA Teaching and Research Center, Gembloux Agro-Bio Tech, University of Liege, Liège, Belgium
- 4Water-Soil-Plant Exchanges Unit, TERRA Teaching and Research Centre, Gembloux Agro-Bio Tech, University of Liege, Liège, Belgium
- 5Forest Soils and Biogeochemistry, Research Institute for Forest, Snow and Landscape Research WSL, Birmensdorf, Switzerland
- 6Sustainable Agroecosystems, Department of Environmental Systems Science, Institute of Agricultural Sciences, Swiss Federal Institute of Technology ETH Zurich, Zurich, Switzerland
Conservation agriculture is based on harnessing and optimizing natural ecological processes such as those mediated by the soil microbiota and represents a very promising approach to overcome the limitations of current conventional practices. Agronomic conservation strategies such as increasing plant diversity at field scale and minimizing soil disturbances such as tillage aim at fostering the presence of beneficial microbial communities to support intrinsic agroecosystem functions and reduce the reliance on agrochemicals and mechanical soil management. However, it remains poorly understood how these positive contributions are modulated by the pedological context. Moreover, only few studies have assessed the management-dependent responses of microbial communities in real-life farming systems. Here, in close association with the farmers and under realistic field conditions, we explored the response of bacterial and fungal community structure to conventional and conservation (using agroecological principles) management regimes across two pedological sites (loamy sand and sandy loam) in Belgium. Microbial diversity was assessed using a DNA metabarcoding approach of ribosomal markers with the Illumina MiSeq sequencing technology. Our results show that different management approaches select for distinct soil microbial communities and that these management-related effects were modulated by the pedological context. Therefore, designing new agricultural systems to foster the presence of beneficial organisms and reduce the level of pathogenic organisms should account for variability in the underlying pedological context.
Introduction
Over the last few decades, several major environmental, economic and societal issues have emerged in relation to agriculture, leading politics, farmers, and scientists to re-think the way we produce food by designing more sustainable agricultural systems (Tilman et al., 2001, 2011). One of the most promising approaches is to design agricultural strategies that mostly rely on biological and naturally occurring ecosystem functions rather than agrochemicals to sustain the productivity of our agroecosystems, while preserving soil resources and reducing the negative impact on environmental and human health (Swinton et al., 2007; Hobbs et al., 2008; Lahmar, 2010; Bender et al., 2016). This approach is not new but it has gained more interest in the last few years as evidenced by the increasing literature containing key words such as “agroecology,” “biodiversity-based agriculture,” “conservation agriculture,” “ecological intensification,” or “organic farming” (Garibaldi et al., 2016; Altieri et al., 2017; Duchene et al., 2017). Basically, the approach of agroecology is based on safeguarding ecological processes and functions that in turn support ecosystem services including crop production (e.g., soil nutrients cycles, pest control) and other ecosystem services that benefit to the society (e.g., aesthetic landscapes, healthy food) (Zhang et al., 2007; Malézieux, 2012; Duru et al., 2015). Even if the concept of agroecology also encompasses other dimensions of food systems such as social and economic components (Francis et al., 2003) and can be defined as a science, a movement and/or a practice (Wezel et al., 2009), we primarily focus here on the practice component. One part of this concept aims at selecting practices that shape the soil biota in a direction that promotes their beneficial services to the ecosystem. Agroecological regimes embrace a wide range of practices such as integrating natural and semi-natural landscape elements, implementing cover crops, using green manure, relying on intercropping or agroforestry, and others (Wezel et al., 2014; Hatt et al., 2016). Numerous studies have identified beneficial effects of these practices to stimulate ecological key functions such as pest regulation by using flower strips (Tschumi et al., 2016) or nutrient acquisition and weed suppression by using intercropping and crop rotations (Liebman and Dyck, 1993; Peoples et al., 2009; Anderson, 2015; Duchene et al., 2017).
In parallel, with the rapid advances in DNA sequencing technologies, we are now able to gain insights into the complex network of below-ground biodiversity (Orgiazzi et al., 2015). Much research in recent years has focused on bacterial and fungal diversity because most of the biological regulations related to organic matter degradation (Ruiz-Dueñas and Martínez, 2009; Trivedi et al., 2016), nutrient acquisition and stimulation of plant growth (Egamberdiyeva, 2007; Philippot et al., 2013; Pii et al., 2015), and supporting plant protection against pests and pathogens (Haas and Défago, 2005) are largely mediated by these groups of microorganisms. Current research activities are now focusing on better understanding and managing below-ground biodiversity through various agricultural strategies in order to promote beneficial microorganisms (Lemanceau et al., 2014; Bender et al., 2016; Delgado-Baquerizo et al., 2016). For example, tillage is now widely recognized as a strong driver of microbial community composition through its global effect on soil properties (Dorr de Quadros et al., 2012; Souza et al., 2013; Säle et al., 2015; Sengupta and Dick, 2015; Tatti et al., 2015; Degrune et al., 2016). However, the limitations and potentials of using agricultural management strategies to engineer the soil microbiota is not well-understood and largely depends on the environmental context (climate and pedological context) varying strongly across the world (Pittelkow et al., 2014).
Although large efforts have been made to build a new biodiversity-based model of agriculture, the contribution of farmers to this knowledge is still underestimated. Most research is done in highly controlled systems where only a few factors vary. Basic scientific studies commonly use simplified factorial systems in order to study isolated components (e.g., tillage, fertilization, etc.) and identify cause-effect relationships. Such simplified systems are able to resolve specific questions by disentangling the individual factors (e.g., Jangid et al., 2008; Ceja-Navarro et al., 2010; Borriello et al., 2012; Dorr de Quadros et al., 2012; Liu et al., 2015; Sengupta and Dick, 2015; Degrune et al., 2016; Danczak et al., 2017). However, in a realistic scenario as faced by farmers in their daily life, these simplified designs are incomplete since they cannot predict the actual performance of real-life systems (Drinkwater, 2002). In real-life systems, agricultural management is the result of choices made by farmers fitting specific economic, social and environmental contexts, and many factors vary simultaneously. As a consequence, testing hypotheses under more realistic conditions has the potential to provide a more holistic understanding of the tradeoffs and synergies among different ecosystem services that occur across environmental, social, and economic dimensions (Kremen and Miles, 2012; Robertson et al., 2014; Ponisio and Kremen, 2016). This study contributes to filling this knowledge gap by exploring the response of bacterial and fungal diversity in agroecological farming systems (AECO) in comparison to conventional farming systems (CONV) using real-life farmer enterprises. The studied AECO and CONV systems are located in the Western Part of the Hainaut Province in Belgium. The AECO systems take part in a self-organizing network of farmers who work together to reach more resilience and autonomy on their farms. Among these, we have selected three cereal fields, which we consider as agroecological as they combine multiple ecological practices: they are organic, implement reduced soil tillage, use intercropping practices and green infrastructures within the farm's landscape. These AECO systems are unique examples of cereal cropping systems with a relatively high level of “agroecologization” as they combine multiple agroecological practices (Horlings and Marsden, 2011; Wezel et al., 2014). Conventional systems are conventionally managed, i.e., applying mineral fertilizers and synthetic weed and pest controls, and using short crop rotations (typical wallonian rotation: winter wheat—beatroot—maize). Selected CONV fields are representative of Walloon cereal farms, while selected AECO fields are “niche examples” of cereal agroecological farms.
The purpose of this study is to describe and examine how agroecological (AECO) management can modulate the response of microbial communities under two different pedological conditions, in comparison with conventional management (CONV). For that purpose, and in close connection with the resident farmers, we have selected six paired-fields in Western Belgium located in two different pedological contexts (loamy sand and sandy loam), though sufficiently close to share similar climatic conditions. A range of physicochemical soil properties were measured and related to data on soil microbial diversity assessed by using a high-throughput DNA sequencing approach of bacterial and fungal ribosomal markers.
Materials and Methods
Experimental Design
The two pedological contexts (sites thereafter) are located in the Western part of the Hainaut province in Belgium (Supplementary Figure 1). The climate is oceanic temperate with an annual rainfall of around 800 mm year−1 and average temperature of around 10°C. The two sites are located in two different geographical regions at Wiers (N50°30′30.122″, E3°31′57.984″) and Gaurain-Ramecroix (N50°35′37.927″, E3°29′21.439″) and geographically separated by about 12 km. At Wiers, the soil is a loamy sand (sandy) and at Gaurain-Ramecroix, the soil is a sandy loam (silty). A preliminary analysis was done using the Soil Map of Wallonia (Bock et al., 2008) in order to select six paired-fields (i.e., three at each site, see Supplementary Figure 1), each pair being homogenous in terms of soil texture, drainage, and soil profile development. This approach aims at removing much of the variation caused by different edaphic properties. The paired sites at the silty site were all characterized as sandy loams, with moderate drainage, and a profile development with a textural Bt horizon. The paired sites at the sandy site were characterized as the following: B1/C1 and B2/C2 are loamy sands, with good drainage and profile development with a textural Bt horizon, whereas B4/C4 are characterized as loamy sands (but softer), with moderate drainage and profile development with a textural Bt horizon. For each pair, a conventionally managed field was paired with a near agroecologically managed field. Finally, in situ soil observation was done to validate the information provided by the Soil Map of Wallonia.
The agroecological fields have been progressively transitioning from conventional (CONV) to agroecological (AECO) management since 1995 (sandy site) and 2010 (silty site). The time frame for the transition from CONV to AECO significantly differed between the sandy and silty site. At the sandy site, plowing was replaced by reduced tillage in 1995 and then by direct seeding in 2010, whereas the use of external fertilizers and pesticides was progressively reduced over time until 2011 when the farm obtained the official Belgian organic certification. At the silty site, however, the transition happened over a shorter time scale, stopping the application of synthetic fertilizers and pesticides and starting organic fertilization in 2011, and subsequently replacing conventional plowing by reduced tillage in 2013. Today, the agroecological management practices at the two sites share similar farming practices: (1) they are certified organic, i.e., no application of agrochemicals through the certification scheme of European and Belgian organic agriculture, (2) rely on reduced tillage and direct seeding, and (3) grow cereals (wheat in the present study) in intercropping procedures and implement green infrastructures (hedgerows, wildflower strips, etc.). The intercropping consists of mixes with the following: triticale, oats, rye, spelt, pea, and vetch. The conventional fields were under wheat conventionally managed, i.e., applying mineral fertilizers and synthetic weed and pest controls, and using short crop rotation (typical local rotation: winter wheat—beetroot—maize). The detailed management of conventional and agroecological fields is fully described in the supplementary material (Supplementary Table 1; agroecological and Supplementary Table 2; conventional management).
Sampling Design and Physico-Chemical Analysis
Soil samples were collected once in July 2015 and all on the same day, a few weeks before harvesting. Within each of the CONV and AECO fields, three plots were randomly identified with GPS points, and within each of these plots, one composite soil sample being a pool of five sub-samples was collected. An auger of 20 cm length and 5 cm width was used to collect the 36 soil samples.
Water content was measured by weighing the sample before and after drying at 105°C according to ISO 11465 (1993). Total C and total N contents were measured by dry combustion (ISO 10694, 1995) under the combined action of elevated temperature (more than 900°C) and oxygen flow. Given the absence of carbonates in the soil samples, the total C measured corresponds to total organic C. Bioavailable elements (Ca, Mg, K, P, and Na) were extracted with ammonium acetate-EDTA 1M (pH 4.65; Lakanen and Erviö, 1971) and quantified by atomic absorption spectroscopy for Ca, Mg, K, and Na and by spectrophotometry for P. Cation-exchange capacity (CEC) was measured using the cobaltihexamine chloride method according to ISO 23470 (2007). Determination of pH(water) and pH(KCl) were carried out by using a soil/solution (water or KCl 1M) ratio of 1:5 (ISO 10390, 2005). Particle size distribution was determined by sedimentation using the pipette method, on three pooled soil samples for each field.
Micro- and Macro-Aggregates Determination
Data for macro- and micro-aggregates, clay, silt, sand and silt-clay content, and aggregate stability were measured as bulk parameter for each field and not determined at the sample level. Aggregate-size separations were conducted on the dry bulk soil by a wet-sieving method adapted from previously published protocols (Elliott, 1986; Six et al., 1998, 2000a). The separation was carried out on three pooled soil samples for each field. Briefly, 80 g of bulk soil sieved at 2 mm was placed on a 250 μm sieve submerged in deionized water for 5 min, at room temperature. Soil was then sieved by moving the sieve out of the deionized water and immersing it again 50 times for 2 min. The same process was repeated again with a 50 μm sieve in order to obtain three fractions: (1) macroaggregates (250–2000 μm), (2) microaggregates (50–250 μm), and (3) silt and clay (<50 μm). All fractions were then dried in an oven at 60°C and weighed in order to determine the mass of each aggregate size fraction.
High-Throughput Sequencing of Bacterial and Fungal Ribosomal Markers
DNA was isolated from 8 g of fresh soil using the PowerMax® soil DNA isolation kit (MO BIO Laboratories, Solana Beach, CA) according to the manufacturer's recommendations. PCR amplification of the bacterial (V3-V4 region of the bacterial 16S rRNA gene) and fungal (ITS2 region of the ribosomal RNA operon) markers was performed as described by Frey et al. (2016). PCR amplification was performed in three technical triplicates and products were pooled prior to sequencing. PCR products were sent to the Génome Québec Innovation Center at McGill University (Montréal, Canada) for barcoding using the Fluidigm Access Array technology (Fluidigm, South San Francisco, CA, USA) and paired-end sequencing on the Illumina MiSeq v3 platform (Illumina Inc., San Diego, CA, USA). The sequence data were processed using a customized pipeline largely based on USEARCH v.11 (Edgar, 2010). In brief, steps included paired-end read merging using USESARCH fastq_mergepairs (Edgar and Flyvbjerg, 2015), filtering PhiX control reads using USEARCH filter_phix, primer trimming using CUTADAPT allowing for one mismatch (Martin, 2011), error filtering by maximum expected error of one using USEARCH fastq_filter (Edgar and Flyvbjerg, 2015), operational taxonomic unit (OTU) delineation (at 97% identity), on-the-fly de novo chimera removal, and singleton removal using USEARCH cluster_otus (Edgar, 2013), as well as target verification using METAXA2 and ITSx (Bengtsson-Palme et al., 2013, 2015). Quality-filtered reads were mapped against the OTU centroid sequences using USEARCH otutab (maxrejects 0, maxaccepts 0, top_hit_only). For taxonomic assignment, OTU centroid sequences were queried against SILVA (Pruesse et al., 2007) and UNITE (Abarenkov et al., 2010) using the SINTAX classifier (Edgar, 2016, 2018) and a minimum bootstrap support of 80%. Non-robust OTUs occurring in <25% of all samples were removed; however, a preliminary analysis revealed similar results between data including all OTUs and only OTUs that occur in at least 25% of all samples.
Statistical Analysis
All statistical analyses were performed using the R software (R Development Core Team, 2018) and Primer6 (Clarke and Gorley, 2006). When required, adjustments for multiple testing were performed using the Benjamini-Hochberg (BH) method (Benjamini and Hochberg, 1995) with the p.adjust function implemented in the vegan package (Oksanen et al., 2007). Differences in β-diversity were examined using Bray-Curtis dissimilarities calculated from standardized (i.e., proportions) and square-root transformed OTU abundances (Hellinger transformation). The significance of the experimental factors was tested using the PERMANOVA routine as implemented in Primer6 (McArdle and Anderson, 2001) with 99,999 permutations. Factors site (2), management type (2) and paired-fields (three paired-fields nested within site) were considered as fixed. The heterogeneity of variance between groups was tested using the PERMDISP routine as implemented in Primer6 with 99,999 permutations. The major variance components of bacterial and fungal β-diversity, as well as the soil physico-chemical parameters were visualized using principal coordinate analyses (PCO) with the cmdscale function Gower (1966) in R. Estimates of α-diversity, i.e., observed richness (Sobs) and Smith-Wilson evenness (Smith and Wilson, 1996), were based on evenly rarefied OTU abundance matrices using an iterative subsampling procedure with 1,000 iterations as implemented in Schloss et al. (2009). Differences in α-diversity and soil physico-chemical parameters were examined using Euclidean distances calculated from z-transformed values and the significance of the experimental factors was tested using the PERMANOVA routine implemented in Primer6 with 99,999 permutations. The relationship between soil properties and β-diversity was assessed using distance-based linear modeling (DistLM, McArdle and Anderson, 2001) implemented in Primer6 with 99,999 permutations. The collinearity among the selected explanatory variables was tested by calculating the variable inflation factor (VIF) using the vif.cca function from vegan. The resulting relationship between soil properties and β-diversity was visualized with distance-based redundancy analysis (dbRDA) using the capscale function from vegan. The correlation between the physico-chemical Euclidean distance matrix and the β-diversity Bray-Curtis distance matrix was assessed by Spearman correlation using the mantel function in vegan. The management-related response of individual taxa was evaluated using univariate permutational analysis of variance (adonis function) based on Euclidean distances calculated from OTU abundances as implemented in vegan package with 99,999 permutations. In order to visualize positive or negative responses of the individual taxa to one of the management types, the relative abundances were z-transformed. The same analysis was performed on the individual soil physico-chemical parameters.
Results
Soil Physical and Chemical Properties
Overall, the pedological context was the main factor discriminating our samples based on the physical and chemical soil properties with the silty site characterized as more humid, nutrient rich, and less acidic than the sandy site (Figures 1A,B). As predicted by the Soil Map of Wallonia, the CONV and AECO fields at the sandy site showed similar clay, silt and sand fractions (Table 1). However, in contrast to our predictions, the two management types have significantly different soil textures at the silty site, with the AECO fields being richer in clay and silt than the CONV fields. At the silty site, one AECO field was very different from the two other AECO fields (Figure 1A). Since soil texture is a known driver of microbial community composition, this difference potentially cofounds any management effect observed at the silty site (see discussion). Differences in the other physicochemical soil properties between CONV and AECO were also often site-dependent (Table 1 and Figure 1C). The management-related response of almost all the descriptors showed discrepancies across the two sites. For example, soil moisture increased under AECO at the sandy site, but was similar between AECO and CONV at the silty site (Figure 1C, moisture). For some parameters, even opposing shifts were observed, e.g., more macro-aggregates under CONV at the sandy site, but more macro-aggregates under AECO at the the silty site (Figure 1C, macro.agg). Furthermore, effects of management on soil properties were not only site dependent, but also paired-field dependent (Supplementary Table 3). The values of the soil variables measured on the 36 samples are provided in Supplementary Table 4.
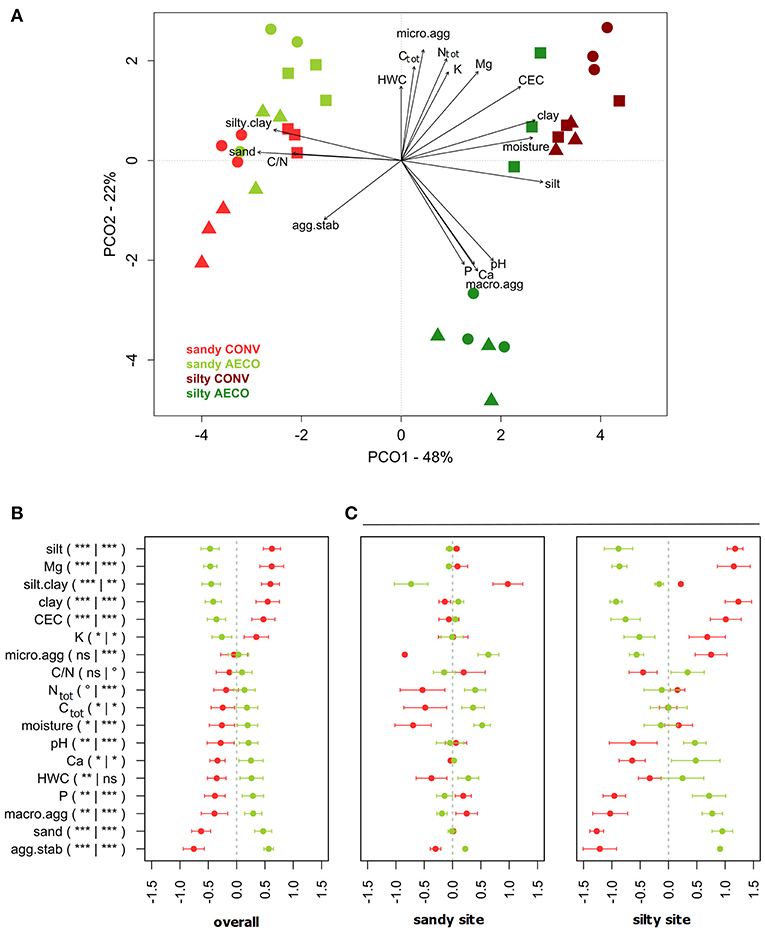
Figure 1. (A) Principal coordinate analysis (PCO) based on Euclidean distances calculated from z-transformed values of physical and chemical soil parameters. The variance explained by the axes (goodness of fit) is provided next to the axis headers. Agricultural management types are colored coded with red (conventional, CONV) and green (agroecological, AECO). Light colors represent the sandy site; dark colors represent the silty site. The symbols represent the pairs: squares, samples belonging to pair n°1; circles, pair n°2; triangles, pair n°3 (see Supplementary Figure 1). (B) Standardized relative change in physical and chemical soil properties calculated from z-transformed values between conventional (red) and agroecological (green) management across all textural sites. (C) Standardized relative change in physical and chemical soil properties calculated from z-transformed values between conventional (red) and agroecological (green) farming systems separately for each textural site. P, K, Mg, and Ca, exchangeable cations; agg.stab, aggregate stability; Ctot, carbon total; clay, % of clay particles; silt, % of silt particles; sand, % of sand particles; micro, % of micro-aggregates; macro, % of macro-aggregates; silt.clay, % of silt and clay fraction; C/N, carbon to nitrogen ratio; HWC, hot water carbon; Ntot, total nitrogen; CEC, cation exchange capacity.
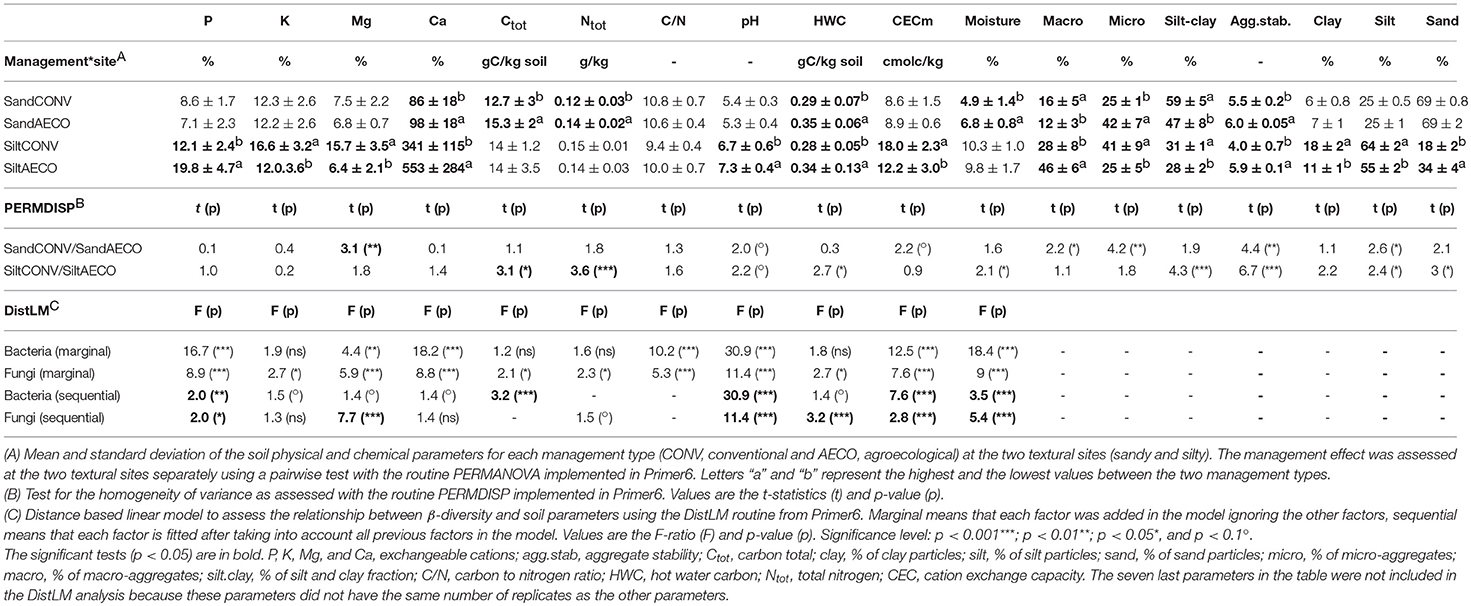
Table 1. Descriptive statistics, test for variance heterogeneity of physical and chemical parameters, and distance linear model of the relationship between physical and chemical parameters and β-diversity.
Microbial Community Structure
Amplicon sequencing yielded a total of 1 267 189 bacterial 16S rRNA gene (35 199 ± 4 922 per sample) and 2 786 747 fungal ITS2 (77 410 ± 15 482 per sample) high-quality sequences from the 36 samples. Sequence clustering yielded a total of 9,583 (3,788 ± 354 per sample) bacterial and 3,032 (820 ± 97 per sample) fungal OTUs. Among the major groups of bacteria, the dominant phyla were Proteobacteria (21%), Acidobacteria (14%), Planctomycetes (9.4%), Verrucomicrobia (9.4%), Actinobacteria (4.5%), and Bacteroidetes (4.4%). The dominant fungal phyla were Ascomycota (44%) and Basidiomycota (5.4%).
The pedological context emerged as the main driver shaping microbial community structure (Figure 2A), explaining 29 and 27% of the variation observed in the bacterial and fungal β-diversity, respectively (Table 2A). Management type was also a significant determinant of the community structure, explaining 10 and 22% of the β-diversity variation in the two datasets. Thus, whereas bacteria appeared to be mainly driven by the sites with a smaller response to the management, fungi appeared to be equally driven by the two factors. In agreement with the shifts in physicochemical properties, the management effects on the microbial communities were site-dependent (Table 2A, management x site), showing somewhat stronger compositional shifts between CONV and AECO at the silty site when compared to the sandy site (Figure 2A). Furthermore, and again equivalent to the physicochemical properties, there was also considerable variation among the paired-fields within each site and management effects were not only site but also paired-field dependent (Table 2, management x paired-field). As revealed by the mantel test, the bacterial (r = 0.51, p = 0.001) and fungal (r = 0.50, p = 0.001) community pattern (Figure 2A) correlated significantly with the physico-chemical pattern (Figure 1A). In a similar way, the bacterial community pattern correlated significantly with the fungal community pattern (r = 0.77, p = 0.001).
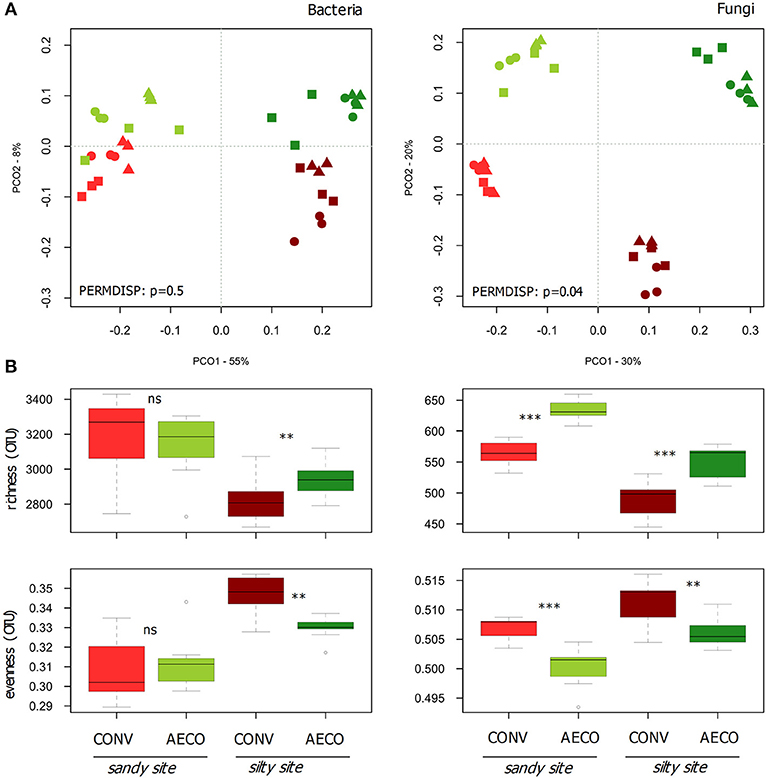
Figure 2. Effects of site and agricultural management on bacterial and fungal β-diversity (A) and α-diversity (B). (A) Principal coordinate analysis (PCO) based on Bray-Curtis similarities calculated from bacterial and fungal OTU abundance matrices. The variance explained by the axes (goodness of fit) is provided next to the axis headers. Agricultural management types are colored coded with red (conventional, CONV) and green (agroecological, AECO). Light colors represent the sandy site; dark colors represent the silty site. Statistical tests for assessing the heterogeneity of variance for the interaction effect between management and site as assessed by PERMDISP are provided in the ordinations. (B) Boxplots of observed richness and Smith-Wilson evenness calculated from the bacterial and fungal OTU matrices. The significant changes are labeled by asterisks: ***p < 0.001, **p < 0.01, °p < 0.1, and ns, non-significant. The p-values were corrected for multiple comparisons using the Benjamini-Hochberg procedure.
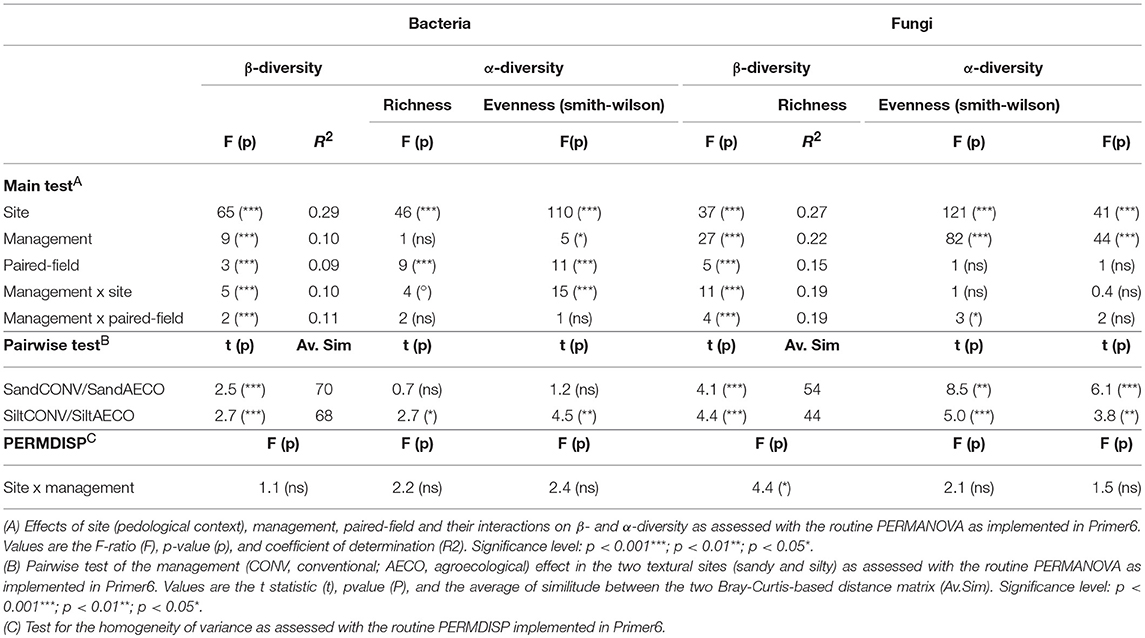
Table 2. Effects of site, management and paired-field on bacterial and fungal β- and α-diversity as assessed by permutational analysis of variance.
Shifts in diversity were also observed at the level of α-diversity (Figure 2B). Bacterial communities showed higher richness and lower evenness under AECO at the silty site, whereas these metrics were similar between AECO and CONV at the sandy site. Fungi showed a more consistent response, being more rich and less even under AECO at both sites. The differences in these parameters were not only depending on the pedological context, but again also on the paired-fields within each pedological site (Supplementary Table 5).
Response of Individual Microbial Taxa to Management
Across both sites, abundant (i.e., ≥1%) bacterial phyla including Gemmatimonadetes, Bacteroidetes, Chloroflexi, and Actinobacteria increased in relative abundance under CONV, whereas Planctomycetes and Acidobacteria increased under AECO (Figure 3A). Likewise, for fungi, Ascomycota increased under CONV, whereas Basidiomycota, Chytridiomycota, and Glomeromycota increased under AECO. However, there can be considerable response heterogeneity within this higher-order groups, evidenced by the fact that Alpha-Proteobacteria increased under CONV and Delta-Proteobacteria increased under AECO. For fungi, the major classes of Ascomycota also did not reveal a uniform response to management type with Sordariomycetes and Pezizomycetes increasing under CONV and Dothideomycetes increasing under AECO. The same was true for the major classes of Basidiomycota with Microbotryomycetes increasing under CONV and Agaricomycetes increasing under AECO. This suggests that (1) management type has shaped the response of individual taxonomic groups and (2) management-related responses were not consistent across the different clades of the major groups.
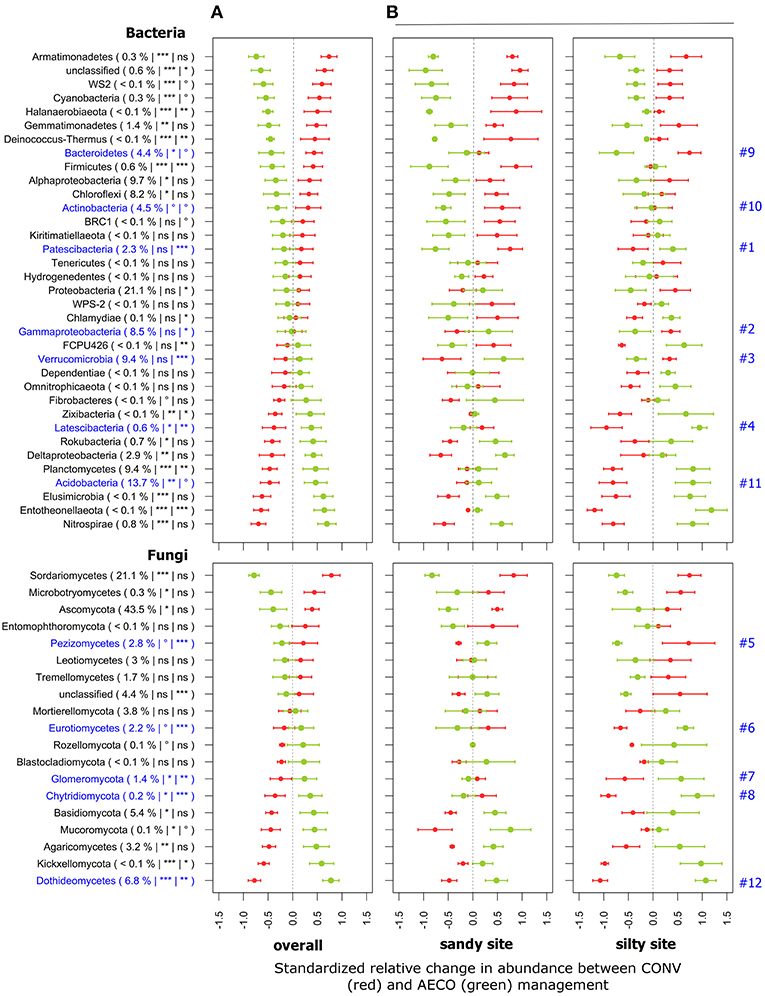
Figure 3. Standardized relative changes in abundance of higher-order taxonomic groups between conventional (red) and agroecological (green) management across all textural sites (A) and separately for each individual textural site (B). Proteobacteria (Alpha-, Gamma-, Delta-proteobacteria), Ascomycota and Basidiomycota were additionally split into the major classes. Data were z-transformed, representing values greater or smaller than the average across all samples. The standardized relative mean and standard deviations are provided for each taxon. The relative abundance as well as the significance of the PERMANOVA test is indicated in brackets: the first argument represents the relative abundance, the second the significance of agricultural management, and the third the significance of the interaction between management and textural site; ***p < 0.001, **p < 0.01, *p < 0.05, °p < 0.1 and ns, not significant. The p-values were corrected for multiple comparisons using the Benjamini-Hochberg procedure. Taxa labeled in blue have revealed a contrasting response to agricultural management across the textural sites as detailed in the main text.
These taxon-level responses often differed between the sites (Figure 3B). Some groups such as Patescibacteria (Figure 3B, #1), Gamma-Proteobacteria (#2), Verrucomicrobia (#3), Latescibacteria (#4), Pezizomycetes (#5), Eurotiomycetes (#6), Glomeromycota (#7), and Chitridiomycota (#8) showed an opposite management-related response between the sites. Other groups such as Bacteroidetes (#9), Actinobacteria (#10), Acidobacteria (#11), and Dothideomycetes (#12) showed a response in the same direction but with different intensity. Whereas, Bacteroidetes (#9), Acidobacteria (#11), and Dothideomycetes (#12) showed a more pronounced response at the silty site, only Actinobacteria (#10) showed a more pronounced response at the sandy site. Again, these taxon-level responses did not only differ between the sites but also between the different paired-fields within each site (Supplementary Figure 2).
Relationship Between Soil Properties and Microbial Community Structure
Distance-based linear modeling using a step-wise selection procedure was used to find a set of physicochemical properties that best predict the observed changes in microbial community structure (Table 1C, sequential test). For bacteria, the most parsimonious sequential model included P, Ctot, pH, CEC, and moisture, explaining a total of 67% of the variation in community structure. For fungi, the most parsimonious model included P, Mg, pH, HWC, CEC, and moisture, explaining a total of 60% of the variation. Overall, pH was the best predictor to explain both bacterial and fungal β-diversity (Table 1C, sequential test). In a second step, distance-based redundancy analysis (db-RDA) was used to visualize the influence of the physicochemical predictors on the bacterial and fungal community structure (Figure 4). The fact that differences in bacterial and fungal community structures as observed in the unconstrained PCO (Figure 2A) were largely recovered in the constrained db-RDA (Figure 4) provides another indication of the substantial effect of the measured physicochemical properties on the microbial community structure. The VIF values of the best predictors of microbial community structure were all below 10 (and all but one below five): bacterial model; P (VIF = 2.88), Ctot (1.32), pH (3.97), CEC (3.24) and moisture (4.46), and fungal model; P (2.96), Mg (4.71), pH (4.53), HWC (1.41), CEC (7.96) and moisture (4.44). We reasonably assume that none of these variables are redundant and that the regression coefficient of the two models is not excessively inflated due to multicollinearity in the model (James et al., 2013).
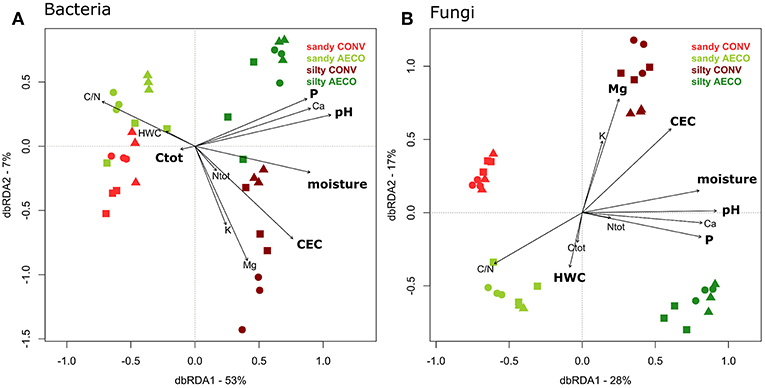
Figure 4. Distance-based redundancy analysis representing the relationship between change in soil properties (represented by the arrows) and (A) bacterial and (B) fungal β-diversity. The variance explained by the axes is given in parentheses and represent the variance explained by all the physical and chemical parameters provided by the arrows. Agricultural management types are colored coded with red (conventional, CONV) and green (agroecological, AECO). Light colors represent the sandy site; dark colors represent the silty site. The symbols represent the pairs: squares, samples belonging to pair n°1; circles, pair n°2; triangles, pair n°3 (see Supplementary Figure 1). The parameters in bold represent the best descriptors as assessed by the DistLM provided in Table 1C.
Discussion
The Pedological Context Matters
This study demonstrates that the pedological context modulates, to some extent, the response of soil physicochemical properties to agricultural management, which will have, in turn, direct implications for the diversity of soil bacterial and fungal communities. In our study, the transition from CONV to AECO management has generally been beneficial for soil quality, increasing soil moisture, stability of soil aggregates, and macro-aggregates. Nutrient availability and pH also increased under AECO management, which might result in better nutrient root uptake, increased microbial activity, and enhanced organic matter degradation. Increasing soil quality through conservational soil management can potentially mitigate some major environmental issues such as soil erosion and soil carbon depletion, and thus improve the functioning. For example, no-till systems have shown to moderate soil erosion (Montgomery, 2007) and decrease turnover of macro-aggregates (Six et al., 2000b), ultimately leading to enhanced carbon protection within macro-aggregates. In addition, a recent study comparing different ecosystem services delivered by the two same systems (AECO and CONV) showed that overall soil aggregate stability and soil respiration rates were more supported by AECO management, whereas higher net crop yields and higher fodder quality were showed under CONV management (Boeraeve et al., submitted). Such results highlight the benefits provided by agroecological management to support various ecosystem services in contrast to focusing only on economic performance.
However, even though the overall soil quality increased under AECO management, we also noticed variation in soil properties between CONV and AECO across the two sites (Figure 1C). In general, changes between CONV and AECO were more pronounced at the silty site, whereas these changes were more moderate at the sandy site. This is an interesting observation given that the transition from CONV to AECO was more recent at the silty than at the sandy site and future investigations should also systematically assess the temporal aspect of the transition from CONV to AECO. Strikingly, numerous key soil parameters showed an opposite response to the soil management (macro- and micro-aggregates, C/N ratio, total nitrogen, phosphorus, and moisture). This site-dependent effect on how CONV and AECO shape the soil conditions will have substantial consequences on the structure of microbial communities and associated functions and processes, which in turn can again change the physical, chemical, and biological conditions of the soil.
Microorganisms living in soil are strongly influenced by their surrounding and changes in soil properties have an impact on their diversity. Our results showed that soil pH was the best predictor of both bacterial and fungal diversity (Table 1). Although previous studies support this evidence for bacteria (Lauber et al., 2008, 2009; Rousk et al., 2010), other drivers including P and N availability were reported to best explain fungal diversity (Lauber et al., 2008; Leff et al., 2015). Besides pH, soil moisture also appeared to be a strong factor shaping both bacterial and fungal community diversity, which is in agreement with previous studies (Fierer et al., 2003; Brockett et al., 2012). In analogy, management-induced changes in major groups of bacteria and fungi were also site-dependent (Figure 3B). Among them, some are known to be potentially beneficial for plant growth and health. For example, arbuscular mycorrhizal fungi (AMF, Glomeromycota), a plant-symbiotic fungi of ecological importance because of its role in plant nutrition and drought tolerance (van der Heijden et al., 2008) generally increased in relative abundance under AECO (Figure 3A, #7), which is in agreement with previous studies highlighting the beneficial effect of conservation agriculture on AMF (Verbruggen et al., 2010). However, our study showed that AECO promoted Glomeromycota only at the silty but not at the sandy site. Similarly, Eurotiomycetes also increased under AECO at the silty site but tended to decrease at the sandy site (Figure 3B, #6). Recent work has shown that Eurotiomycetes can potentially degrade moderately labile and recalcitrant forms of carbon better than other groups of fungi by producing higher levels of Xylosidases and Glucosaminidases (Trivedi et al., 2016). Among bacteria, Acidobacteria, a group of largely oligotrophic organisms (Fierer et al., 2007) that are known to be sensitive to pH (Rousk et al., 2010), also tended to increase under AECO at the silty but not much at the sandy site (Figure 3B, #11), as did the soil pH (Figure 1C). Although fungi are generally considered as the major microbial decomposers of plant materials, previous studies showed a clear enrichment of acid-tolerant Acidobacteria under lignin-amended conditions which suggests a putative role of this bacterial group in degradation of complex organic material (DeAngelis et al., 2011).
Two less well-known groups of bacteria, the Patescibacteria (Figure 3, #1) and Latescibacteria (#4) also showed strong soil-type dependant responses. These two groups increased under AECO at the silty site, but tended to decrease at the sandy site. These two groups belong to yet-uncultured microbial candidate phyla and therefore their metabolic capabilities and ecological roles are not yet well-understood. Recent studies have highlighted the prevalent saprophytic lifestyle in the Latescibacteria lineage (member of the Fibrobacteres-Chlorobi-Bacteroidetes (FCB) superphylum) suggesting their important role in soil organic detritus degradation (Farag et al., 2017). Patescibacteria belong to the candidate phyla radiation (CPR) (Brown et al., 2015), a phylogenetic super-clade that is characterized by limited metabolic capabilities, and small streamlined genomes that might point to symbiotic or syntrophic lifestyles (Wrighton et al., 2012; Nelson and Stegen, 2015). Recent work further emphasized the potential role of CPR members in carbon degradation (Danczak et al., 2017). Although the contribution of this group of bacteria to various ecosystems services is not yet understood, we hypothesize based on the aforementioned metabolic capabilities and ecological roles that these organisms might contribute to soil functioning.
The substantial changes in community structure between CONV and AECO (Figures 2A,B) is in agreement with numerous previous studies showing effects of agricultural management on soil microbial community structures by means of changing the soil conditions (Ceja-Navarro et al., 2010; Chaudhry et al., 2012; Hartmann et al., 2015; Degrune et al., 2017; Lupatini et al., 2017). In our study, AECO management showed higher microbial diversity (i.e., richness) when compared to CONV management. Previous studies reported an increase in microbial diversity under organic management (Chaudhry et al., 2012; Hartmann et al., 2015). When assessing the effect of agricultural management on microbial diversity, most studies have focused in the past on the effect of tillage regimes (Ceja-Navarro et al., 2010; Dorr de Quadros et al., 2012; Sengupta and Dick, 2015; Degrune et al., 2016) and very few studies have addressed the question of agroecological practices, and how the combination of several conservation practices may interact with the diversity of the soil microbiota. Although increasing the actual diversity seems a pillar argument to support the transition from conventional to organic or conservation agriculture, we still have a poor understanding of how actual changes in diversity per se translate into changes of the ecological processes mediated by these communities. Thus, an increase in diversity alone is clearly not enough to advocate transition toward agroecological systems. The composition of the community is thereby a central component shaping the functional capacity of the soil.
For example, a key aspect from the farmer's perspective is the potential of a specific management system to mitigate the presence of soil pathogens and/or promote the presence of beneficial species to ultimately better support crop growth and health. With the absence of synthetic pesticides to manage soil pathogens and reduced inputs of mineral fertilizer to secure plant nutrition, such management systems clearly rely on efficient below-ground biotic interactions. Previous studies have suggested that organic or other forms of conservation management have the ability to reduce pressure from pest and pathogens and promote beneficial species important for plant nutrition and immunity (van Bniggen and Termorskuizen, 2003; Birkhofer et al., 2008; Peoples et al., 2009; Bruggen et al., 2016; Tschumi et al., 2016). In contrast to conventional farming systems where pest and disease regulation regimes specifically target the pathogen itself, organic or other forms of conservation management largely rely on indirect mechanisms for plant protection and nutrition. For example, diversifying cropping systems will promote nitrogen-fixing symbiosis in the case of legumes association (Peoples et al., 2009). Numerous other mechanisms under organic management are well-described in Bruggen et al. (2016). In order to draw conclusions about potential pathogenic or beneficial taxa, data need to be interrogated at lower taxonomic levels and we provide data on management effects for all detected OTUs in Supplementary Table 6. For example, OTUs assigned to Fusarium, a genus of potential fungal wheat pathogens (Dean et al., 2012; Perelló and Larrán, 2013), were found to increase both under CONV and AECO management (in this case only two OTUs), whereas Alternaria species, another group of potential fungal pathogens increased under AECO management. Putative beneficial fungi such as Glomus and Funneliformis were substantially increased under AECO management. For bacteria, however, none of the commonly known plant pathogenic taxa were found to differ between the management systems. Such conclusions about potential pathogenic and beneficial taxa need to be taken with caution, however, since accurate identification at lower taxonomic levels is limited by both the short-read length of the technology and the limited resolution of the genetic markers used, making it difficult to make highly accurate species level predictions. Furthermore, an actual pathogenic or beneficial activity of a species cannot be solely deduced from its taxonomic status. Overall, it has to be understood that the multitude of factors that are simultaneously changed under agroecological management approaches induce a series of complex and interdependent effects on the soil microbial network that cannot be quantified by solely looking at the presence or absence of individual beneficial or pathogenic species.
The hypothesized positive relationship between biodiversity and ecosystem functioning have long been a central but still unresolved question in biodiversity research (Coleman and Whitman, 2005; Byrnes et al., 2014; Ricketts et al., 2016). However, evaluating this relationship under real-life scenarios where farmers make specific management decisions that might change over time, make it difficult to predict the output of this relationship. Therefore, even if AECO can promote the presence of beneficial microorganisms (such as arbuscular mycorrhizal fungi) and mitigate pressure from pathogens, we ultimately cannot predict, if it will improve food production or any other ecosystem service. In this light, there is still little evidence that farming systems that promote higher microbial diversity actually increase the performance of the agroecosystem by securing more ecosystem functions and making it less vulnerable to extreme events.
New Insights Through On-Farm System-Level Assessments
Most studies use factorial approaches to break down complex systems like agroecosystems into isolated components in order to identify cause-effect relationships (Drinkwater, 2002). Such approaches enable us to answer important agronomic questions by quantifying the relative importance of individual management factors such as tillage (e.g., plowing, reduced tillage, no tillage), fertilization (e.g., compost, manure, synthetic), and plant diversity (e.g., intercropping, cover cropping, crop rotation) on specific agronomic outcomes such as crop yield, germination rate, soil structure, biological activity, and more recently with the advance in molecular biology the structure of soil microbial communities (Roesch et al., 2007; Delmont et al., 2011; Franzosa et al., 2015; Orgiazzi et al., 2015). These factorial approaches to study cause-effect relationships have, however, also some practical limitations (Drinkwater, 2002) and a more holistic system-level assessment representing a realistic agronomic, social, and economic context can provide additional insights in order to ultimately link knowhow gained from classical research designs with realistic on-farm observations. The more holistic, farm-level approach carried out in this study aims at understanding how the combination of several individual management factors affects the soils in the context of the farmer's dynamic decisions to modify the agronomic outcomes. We fully acknowledge that our approach sacrifices resolution, since the relative importance of each factor cannot be disentangled; but the advantages gained by this approach are measurable outcomes (here, how the structure of microbial communities responds to agricultural management, or in Boeraeve et al. (submitted) the delivery of ecosystem services) that result from the positive or negative synergies occurring among the different factors combined together in a realistic way (i.e., decided by a farmer who takes into account multiple factors when making agricultural decisions). Also, it is worth mentioning that replicates in a holistic farm-level approach do not have the same meaning as replicates used in a classic experimental approach. For example, in our study we know that fields from the same agricultural management are managed by different farmers, each of them making choices based on non-scientific motivations and concerns that are based on social and economic reasons, which may also change over time. Therefore, the actual agricultural management might differ slightly from one field to another even though the philosophy remains the same, and will be highly dependent on a farmer's habits and preferences. At the sandy site for instance, the transition toward AECO management started 20 years ago, was progressive and is still going, with farmer practices changing slowly over time. Plowing was first stopped and then the amount on agrochemical was progressively decreased by 2012 to the point where no agrochemicals were used. At the silty site, the transition began more recently and it is still evolving. As a consequence, AECO-managed fields at the sandy and silty sites were not at the same stage of transition and thus the physical and chemical conditions occurring in the soil at sampling time did not reflect the final conditions as found in stabilized systems. Interestingly though, the effect at the silty site was more pronounced despite its more recent transition, indicating that the soil type might be an important determinant of how rapidly these transition effects establish. Importantly, farming systems are highly complex systems where numerous components interact (e.g., practices, farmer's choices, economic context, pedological context, etc.). Therefore, understanding such complex systems to design more performant and efficient agriculture require the use of not only one approach (holistic vs. factorial), but the use of system-based and multidimensional approaches (Kremen and Miles, 2012; Robertson et al., 2014; Ponisio Lauren et al., 2015).
Overall, our study provided more insights into how the transition to agroecological farming systems initiated by Belgian farmers several years ago can potentially enhance the performance (i.e., ecosystem functions and services) of the system by (1) improving the overall soil quality through different parameters (nutrient status, soil moisture, soil aggregation, etc.) and (2) increasing the relative abundance of beneficial groups of bacteria and fungi. Here, the word “potentially” has its importance since we have no evidence on the relationship of these diversity changes and the actual performance of the system. Even if we had observed an improvement of several soil and microbial biodiversity parameters under AECO management along with a clear shift in the community structure, the question whether Belgian farmers should transition to AECO farming system cannot be solely addressed with soil data. Even if our study provides a small but nevertheless solid brick to address this crucial question, other aspects of the system must be evaluated as well. For example, the financial aspect (e.g., Crowder and Reganold, 2015) must play a major role into the farmer's decision of a transition. The actual knowledge of the farmers themselves must also be considered before deciding on transitioning (Boeraeve et al., 2018; Šūmane et al., 2018). Similarly, the environmental context may as well play a critical role. As we have shown in our study, the management-related response of the soil microbial community was strongly modulated by the pedological context. Therefore, the benefits that the farmers may gain from the transition could be, in the same way, modulated by the pedological context. The question of the transition to agroecological farming systems remains highly complex and requires a more holistic and complete understanding of all the trade-offs of such a transition.
Large efforts are currently made to develop management strategies that enhance soil biodiversity and manipulate the community composition of microbes to target specific beneficial or pathogenic species. By unraveling the response of bacterial and fungal diversity to agroecological management under different pedological context on real-life farms, our study, despite covering a rather narrow gradient of pedological variation and considering only one type of climate, has contributed to provide a first stepping stone to address this knowledge gap. Agroecological management is dynamic and will be continuing evolving. Studying such system will allow to better understand the multiple interactions between management and outcome (here microbial diversity) in order to design innovative and ecologically, economically, and socially beneficial agroecological practices. These findings suggest that such species response patterns must be highly variable across different soil types, calling for large-scale surveys across much broader gradients of these factors in order to get a more complete understanding of the tripartite interaction between soil, management, and microbiome.
Data Availability
The microbial datasets for this study can be found in the European Nucleotide Archive under the accession number PRJEB29246.
Author Contributions
MD and J-TC conceived the original idea. MD wrote the project. FD, FB, and J-TC carried out the experiment. FD and MH performed the bioinformatics and statistical analyses and wrote the manuscript. All authors contributed to the final manuscript, provided critical feedback, and helped shaping the analysis.
Funding
This study was funded by FNRS Crédit de Recherche Farms For Future–F4F and AgricultureIsLife (Gembloux Agro Bio tech) with financial contributions of the Swiss Federal Research Institute WSL.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Joachim Tulii for helping with the experiment. We thank Beat Stierli at the Swiss Federal Research Institute WSL for the laboratory work. We further acknowledge the contribution of the staff at the McGill University and Génome Québec Innovation Center, Montreal, Canada, for the sequencing service. We also thank the Genetic Diversity Center (GDC) at ETH Zürich for providing computational resources.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fevo.2019.00261/full#supplementary-material
References
Abarenkov, K., Nilsson, R. H., Larsson, K.-H., Alexander, H., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi – recent updates and future perspectives. N. Phytol. 186, 281–285. doi: 10.1111/j.1469-8137.2009.03160.x
Altieri, M., Nicholls, C., and Montalba, R. (2017). Technological approaches to sustainable agriculture at a crossroads: an agroecological perspective. Sustainability 9:349. doi: 10.3390/su9030349
Anderson, R. L. (2015). Integrating a complex rotation with no-till improves weed management in organic farming. A review. Agron. Sustain. Dev. 35, 967–974. doi: 10.1007/s13593-015-0292-3
Bender, S. F., Wagg, C., and van der Heijden, M. G. A. (2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 31, 440–452. doi: 10.1016/j.tree.2016.02.016
Bengtsson-Palme, J., Hartmann, M., Eriksson, K. M., Pal, C., Thorell, K., Larsson, D. G. J., et al. (2015). Metaxa2: improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 15, 1403–1414. doi: 10.1111/1755-0998.12399
Bengtsson-Palme, J., Ryberg, M., Hartmann, M., Branco, S., Wang, Z., Godhe, A., et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 4, 914–919. doi: 10.1111/2041-210X.12073
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x
Birkhofer, K., Bezemer, T. M., Bloem, J., Bonkowski, M., Christensen, S., Dubois, D., et al. (2008). Long-term organic farming fosters below and aboveground biota: implications for soil quality, biological control and productivity. Soil Biol. Biochem. 40, 2297–2308. doi: 10.1016/j.soilbio.2008.05.007
Bock, L., Legrain, X., Veron, P., Bracke, C., Bah, B., and Lejeune, P. (2008). Carte Numérique des Sols de Wallonie–Version 1.2, Gembloux.
Boeraeve, F., Dufrene, M., De Vreese, R., Jacobs, S., Pipart, N., Turkelboom, F., et al. (2018). Participatory identification and selection of ecosystem services: building on field experiences. Ecol. Soc. 23:27. doi: 10.5751/ES-10087-230227
Borriello, R., Lumini, E., Girlanda, M., Bonfante, P., and Bianciotto, V. (2012). Effects of different management practices on arbuscular mycorrhizal fungal diversity in maize fields by a molecular approach. Biol. Fertil. Soils 48, 911–922. doi: 10.1007/s00374-012-0683-4
Brockett, B. F. T., Prescott, C. E., and Grayston, S. J. (2012). Soil moisture is the major factor influencing microbial community structure and enzyme activities across seven biogeoclimatic zones in western Canada. Soil Biol. Biochem. 44, 9–20. doi: 10.1016/j.soilbio.2011.09.003
Brown, C. T., Hug, L. A., Thomas, B. C., Sharon, I., Castelle, C. J., Singh, A., et al. (2015). Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523, 208–211. doi: 10.1038/nature14486
Bruggen, A. H., van Gamliel, A., and Finckh, M. R. (2016). Plant disease management in organic farming systems. Pest Manag. Sci. 72, 30–44. doi: 10.1002/ps.4145
Byrnes, J. E. K., Gamfeldt, L., Isbell, F., Lefcheck, J. S., Griffin, J. N., Hector, A., et al. (2014). Investigating the relationship between biodiversity and ecosystem multifunctionality: challenges and solutions. Methods Ecol. Evol. 5, 111–124. doi: 10.1111/2041-210X.12143
Ceja-Navarro, J. A., Rivera-Orduña, F. N., Patiño-Zúñiga, L., Vila-Sanjurjo, A., Crossa, J., Govaerts, B., et al. (2010). Phylogenetic and multivariate analyses to determine the effects of different tillage and residue management practices on soil bacterial communities. Appl. Environ. Microbiol. 76, 3685–3691. doi: 10.1128/AEM.02726-09
Chaudhry, V., Rehman, A., Mishra, A., Chauhan, P. S., and Nautiyal, C. S. (2012). Changes in bacterial community structure of agricultural land due to long-term organic and chemical amendments. Microb. Ecol. 64, 450–460. doi: 10.1007/s00248-012-0025-y
Coleman, D. C., and Whitman, W. B. (2005). Linking species richness, biodiversity and ecosystem function in soil systems. Pedobiologia 49, 479–497. doi: 10.1016/j.pedobi.2005.05.006
Crowder, D. W., and Reganold, J. P. (2015). Financial competitiveness of organic agriculture on a global scale. Proc. Natl. Acad. Sci. 112, 7611–7616. doi: 10.1073/pnas.1423674112
Danczak, R. E., Johnston, M. D., Kenah, C., Slattery, M., Wrighton, K. C., and Wilkins, M. J. (2017). Members of the candidate phyla radiation are functionally differentiated by carbon-and nitrogen-cycling capabilities. Microbiome 5:112. doi: 10.1186/s40168-017-0331-1
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
DeAngelis, K. M., Allgaier, M., Chavarria, Y., Fortney, J. L., Hugenholtz, P., Simmons, B., et al. (2011). Characterization of trapped lignin-degrading microbes in tropical forest soil. PLoS ONE 6:e19306. doi: 10.1371/journal.pone.0019306
Degrune, F., Theodorakopoulos, N., Colinet, G., Hiel, M.-P., Bodson, B., Taminiau, B., et al. (2017). Temporal dynamics of soil microbial communities below the seedbed under two contrasting tillage regimes. Front. Microbiol. 8:1127. doi: 10.3389/fmicb.2017.01127
Degrune, F., Theodorakopoulos, N., Dufrêne, M., Colinet, G., Bodson, B., Hiel, M.-P., et al. (2016). No favorable effect of reduced tillage on microbial community diversity in a silty loam soil (Belgium). Agric. Ecosyst. Environ. 224, 12–21. doi: 10.1016/j.agee.2016.03.017
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7:10541. doi: 10.1038/ncomms10541
Delmont, T. O., Robe, P., Cecillon, S., Clark, I. M., Constancias, F., Simonet, P., et al. (2011). Accessing the soil metagenome for studies of microbial diversity. Appl. Environ. Microbiol. 77, 1315–1324. doi: 10.1128/aem.01526-10
Dorr de Quadros, P., Zhalnina, K., Davis-Richardson, A., Fagen, J. R., Drew, J., Bayer, C., et al. (2012). The effect of tillage system and crop rotation on soil microbial diversity and composition in a subtropical acrisol. Diversity 4, 375–395. doi: 10.3390/d4040375
Drinkwater, L. E. (2002). Cropping systems rsearch: reconsidering agricultural experimental approaches. HortTechnology 12, 355–361. doi: 10.21273/HORTTECH.12.3.355
Duchene, O., Vian, J.-F., and Celette, F. (2017). Intercropping with legume for agroecological cropping systems: complementarity and facilitation processes and the importance of soil microorganisms. A review. Agric. Ecosyst. Environ. 240, 148–161. doi: 10.1016/j.agee.2017.02.019
Duru, M., Therond, O., Martin, G., Martin-Clouaire, R., Magne, M.-A., Justes, E., et al. (2015). How to implement biodiversity-based agriculture to enhance ecosystem services: a review. Agron. Sustain. Dev. 35, 1259–1281. doi: 10.1007/s13593-015-0306-1
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi: 10.1038/nmeth.2604
Edgar, R. C. (2016). SINTAX: a simple non-Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv. doi: 10.1101/074161
Edgar, R. C. (2018). Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ 6:e4652. doi: 10.7717/peerj.4652
Edgar, R. C., and Flyvbjerg, H. (2015). Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 31, 3476–3482. doi: 10.1093/bioinformatics/btv401
Egamberdiyeva, D. (2007). The effect of plant growth promoting bacteria on growth and nutrient uptake of maize in two different soils. Appl. Soil Ecol. 36, 184–189. doi: 10.1016/j.apsoil.2007.02.005
Elliott, E. T. (1986). Aggregate structure and carbon, nitrogen, and phosphorus in native and cultivated soils. Soil Sci. Soc. Am. J. 50, 627–633. doi: 10.2136/sssaj1986.03615995005000030017x
Farag, I. F., Youssef, N. H., and Elshahed, M. S. (2017). Global distribution patterns and pangenomic diversity of the candidate phylum “Latescibacteria”(WS3). Appl Env. Microbiol 83, e00521–e00517. doi: 10.1128/AEM.00521-17
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology 88, 1354–1364. doi: 10.1890/05-1839
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Influence of drying–rewetting frequency on soil bacterial community structure. Microb. Ecol. 45, 63–71. doi: 10.1007/s00248-002-1007-2
Francis, C., Lieblein, G., Gliessman, S., Breland, T. A., Creamer, N., Harwood, R., et al. (2003). Agroecology: the ecology of food systems. J. Sustain. Agric. 22, 99–118. doi: 10.1300/J064v22n03_10
Franzosa, E. A., Hsu, T., Sirota-Madi, A., Shafquat, A., Abu-Ali, G., Morgan, X. C., et al. (2015). Sequencing and beyond: integrating molecular ‘omics' for microbial community profiling. Nat. Rev. Microbiol. 13, 360–372. doi: 10.1038/nrmicro3451
Frey, B., Rime, T., Phillips, M., Stierli, B., Hajdas, I., Widmer, F., et al. (2016). Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 92:fiw018. doi: 10.1093/femsec/fiw018
Garibaldi, L. A., Gemmill-Herren, B., D'Annolfo, R., Graeub, B. E., Cunningham, S. A., and Breeze, T. D. (2016). Farming approaches for greater biodiversity, livelihoods, and food security. Trends Ecol. Evol. 32, 68–80. doi: 10.1016/j.tree.2016.10.001
Gower, J. C. (1966). Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika 53, 325–338. doi: 10.1093/biomet/53.3-4.325
Haas, D., and Défago, G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. doi: 10.1038/nrmicro1129
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hatt, S., Artru, S., Brédart, D., Lassois, L., Francis, F., Haubruge, E., et al. (2016). Towards sustainable food systems: the concept of agroecology and how it questions current research practices. A review. Biotechnol. Agron. Société Environ. 20, 215–224. Available online at: http://hdl.handle.net/2268/197503
Hobbs, P. R., Sayre, K., and Gupta, R. (2008). The role of conservation agriculture in sustainable agriculture. Philos. Trans. R. Soc. B Biol. Sci. 363, 543–555. doi: 10.1098/rstb.2007.2169
Horlings, L. G., and Marsden, T. K. (2011). Towards the real green revolution? Exploring the conceptual dimensions of a new ecological modernisation of agriculture that could ‘feed the world. 'Glob. Environ. Change 21, 441–452. doi: 10.1016/j.gloenvcha.2011.01.004
James, G., Witten, D., Hastie, T., and Tibshirani, R. (2013). An Introduction to Statistical Learning. New York, NY: Springer. doi: 10.1007/978-1-4614-7138-7
Jangid, K., Williams, M. A., Franzluebbers, A. J., Sanderlin, J. S., Reeves, J. H., Jenkins, M. B., et al. (2008). Relative impacts of land-use, management intensity and fertilization upon soil microbial community structure in agricultural systems. Soil Biol. Biochem. 40, 2843–2853. doi: 10.1016/j.soilbio.2008.07.030
Kremen, C., and Miles, A. (2012). Ecosystem services in biologically diversified versus conventional farming systems: benefits, externalities, and trade-offs. Ecol. Soc. 17:40. doi: 10.5751/ES-05035-170440
Lahmar, R. (2010). Adoption of conservation agriculture in Europe: lessons of the KASSA project. Land Use Policy 27, 4–10. doi: 10.1016/j.landusepol.2008.02.001
Lakanen, E., and Erviö, R. (1971). A Comparison of Eight Extractants for the Determination of Plant Available Micronutrients in Soils. Helsinki. Available online at: http://jukuri.luke.fi/handle/10024/472471
Lauber, C. L., Hamady, M., Knight, R., and Fierer, N. (2009). Pyrosequencing-based assessment of soil ph as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 75, 5111–5120. doi: 10.1128/AEM.00335-09
Lauber, C. L., Strickland, M. S., Bradford, M. A., and Fierer, N. (2008). The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 40, 2407–2415. doi: 10.1016/j.soilbio.2008.05.021
Leff, J. W., Jones, S. E., Prober, S. M., Barberán, A., Borer, E. T., Firn, J. L., et al. (2015). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. U.S.A. 112, 10967–10972. doi: 10.1073/pnas.1508382112
Lemanceau, P., Maron, P.-A., Mazurier, S., Mougel, C., Pivato, B., Plassart, P., et al. (2014). Understanding and managing soil biodiversity: a major challenge in agroecology. Agron. Sustain. Dev. 35, 67–81. doi: 10.1007/s13593-014-0247-0
Liebman, M., and Dyck, E. (1993). Crop Rotation and intercropping strategies for weed management. Ecol. Appl. 3, 92–122. doi: 10.2307/1941795
Liu, Y., Johnson, N. C., Mao, L., Shi, G., Jiang, S., Ma, X., et al. (2015). Phylogenetic structure of arbuscular mycorrhizal community shifts in response to increasing soil fertility. Soil Biol. Biochem. 89, 196–205. doi: 10.1016/j.soilbio.2015.07.007
Lupatini, M., Korthals, G. W., de Hollander, M., Janssens, T. K. S., and Kuramae, E. E. (2017). Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 7:2064. doi: 10.3389/fmicb.2016.02064
Malézieux, E. (2012). Designing cropping systems from nature. Agron. Sustain. Dev. 32, 15–29. doi: 10.1007/s13593-011-0027-z
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
McArdle, B. H., and Anderson, M. J. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82, 290–297. doi: 10.1890/0012-9658(2001)082[0290:FMMTCD]2.0.CO;2
Montgomery, D. R. (2007). Soil erosion and agricultural sustainability. Proc. Natl. Acad. Sci. U.S.A. 104, 13268–13272. doi: 10.1073/pnas.0611508104
Nelson, W. C., and Stegen, J. C. (2015). The reduced genomes of Parcubacteria (OD1) contain signatures of a symbiotic lifestyle. Front. Microbiol. 6:713. doi: 10.3389/fmicb.2015.00713
Oksanen, J., Kindt, R., Legendre, P., O'Hara, B., Stevens, M. H. H., Oksanen, M. J., et al. (2007). The vegan package. Community Ecol. Package, 631–637.
Orgiazzi, A., Dunbar, M. B., Panagos, P., de Groot, G. A., and Lemanceau, P. (2015). Soil biodiversity and DNA barcodes: opportunities and challenges. Soil Biol. Biochem. 80, 244–250. doi: 10.1016/j.soilbio.2014.10.014
Peoples, M. B., Brockwell, J., Herridge, D. F., Rochester, I. J., Alves, B. J. R., Urquiaga, S., et al. (2009). The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis 48, 1–17. doi: 10.1007/BF03179980
Perelló, A. E., and Larrán, S. (2013). Nature and effect of Alternaria spp. complex from wheat grain on germination and disease transmission. Pak. J. Bot. 45, 1817–1824.
Philippot, L., Spor, A., Hénault, C., Bru, D., Bizouard, F., Jones, C. M., et al. (2013). Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7, 1609–1619. doi: 10.1038/ismej.2013.34
Pii, Y., Mimmo, T., Tomasi, N., Terzano, R., Cesco, S., and Crecchio, C. (2015). Microbial interactions in the rhizosphere: beneficial influences of plant growth-promoting rhizobacteria on nutrient acquisition process. A review. Biol. Fertil. Soils 51, 403–415. doi: 10.1007/s00374-015-0996-1
Pittelkow, C. M., Liang, X., Linquist, B. A., van Groenigen, K. J., Lee, J., Lundy, M. E., et al. (2014). Productivity limits and potentials of the principles of conservation agriculture. Nature 517, 365–368. doi: 10.1038/nature13809
Ponisio Lauren, C., M'Gonigle Leithen, K., Mace Kevi, C., Palomino, J., de Valpine, P., and Kremen, C. (2015). Diversification practices reduce organic to conventional yield gap. Proc. R. Soc. B, Biol. Sci. 282:20141396. doi: 10.1098/rspb.2014.1396
Ponisio, L. C., and Kremen, C. (2016). System-level approach needed to evaluate the transition to more sustainable agriculture. Proc. R. Soc. B. 283:20152913. doi: 10.1098/rspb.2015.2913
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196. doi: 10.1093/nar/gkm864
R Development Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing Available online at: http://www.R-project.org/
Ricketts, T. H., Watson, K. B., Koh, I., Ellis, A. M., Nicholson, C. C., Posner, S., et al. (2016). Disaggregating the evidence linking biodiversity and ecosystem services. Nat. Commun. 7:13106. doi: 10.1038/ncomms13106
Robertson, G. P., Gross, K. L., Hamilton, S. K., Landis, D. A., Schmidt, T. M., Snapp, S. S., et al. (2014). Farming for ecosystem services: an ecological approach to production agriculture. BioScience 64, 404–415. doi: 10.1093/biosci/biu037
Roesch, L. F. W., Fulthorpe, R. R., Riva, A., Casella, G., Hadwin, A. K. M., Kent, A. D., et al. (2007). Pyrosequencing enumerates and contrasts soil microbial diversity. Isme J. 1:283. doi: 10.1038/ismej.2007.53
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Ruiz-Dueñas, F. J., and Martínez, Á. T. (2009). Microbial degradation of lignin: how a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2, 164–177. doi: 10.1111/j.1751-7915.2008.00078.x
Säle, V., Aguilera, P., Laczko, E., Mäder, P., Berner, A., Zihlmann, U., et al. (2015). Impact of conservation tillage and organic farming on the diversity of arbuscular mycorrhizal fungi. Soil Biol. Biochem. 84, 38–52. doi: 10.1016/j.soilbio.2015.02.005
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541.
Sengupta, A., and Dick, W. A. (2015). Bacterial community diversity in soil under two tillage practices as determined by pyrosequencing. Microb. Ecol. 70, 853–859. doi: 10.1007/s00248-015-0609-4
Six, J., Elliott, E. T., and Paustian, K. (2000a). Soil macroaggregate turnover and microaggregate formation: a mechanism for C sequestration under no-tillage agriculture. Soil Biol. Biochem. 32, 2099–2103. doi: 10.1016/S0038-0717(00)00179-6
Six, J., Elliott, E. T., Paustian, K., and Doran, J. W. (1998). Aggregation and soil organic matter accumulation in cultivated and native grassland soils. Soil Sci. Soc. Am. J. 62:1367. doi: 10.2136/sssaj1998.03615995006200050032x
Six, J., Paustian, K., Elliott, E. T., and Combrink, C. (2000b). Soil structure and organic matter I. Distribution of aggregate-size classes and aggregate-associated carbon. Soil Sci. Soc. Am. J. 64, 681–689. doi: 10.2136/sssaj2000.642681x
Smith, B., and Wilson, J. B. (1996). A consumer's guide to evenness indices. Oikos 76, 70–82. doi: 10.2307/3545749
Souza, R. C., Cantão, M. E., Vasconcelos, A. T. R., Nogueira, M. A., and Hungria, M. (2013). Soil metagenomics reveals differences under conventional and no-tillage with crop rotation or succession. Appl. Soil Ecol. 72, 49–61. doi: 10.1016/j.apsoil.2013.05.021
Šūmane, S., Kunda, I., Knickel, K., Strauss, A., Tisenkopfs, T., Rios, I. d. I., et al. (2018). Local and farmers' knowledge matters! How integrating informal and formal knowledge enhances sustainable and resilient agriculture. J. Rural Stud. 59, 232–241. doi: 10.1016/j.jrurstud.2017.01.020
Swinton, S. M., Lupi, F., Robertson, G. P., and Hamilton, S. K. (2007). Ecosystem services and agriculture: cultivating agricultural ecosystems for diverse benefits. Ecol. Econ. 64, 245–252. doi: 10.1016/j.ecolecon.2007.09.020
Tatti, E., Goyer, C., Burton, D. L., Wertz, S., Zebarth, B. J., Chantigny, M., et al. (2015). Tillage management and seasonal effects on denitrifier community abundance, gene expression and structure over winter. Microb. Ecol. 70, 795–808. doi: 10.1007/s00248-015-0591-x
Tilman, D., Balzer, C., Hill, J., and Befort, B. L. (2011). Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. U.S.A. 108, 20260–20264. doi: 10.1073/pnas.1116437108
Tilman, D., Fargione, J., Wolff, B., D'Antonio, C., Dobson, A., Howarth, R., et al. (2001). Forecasting agriculturally driven global environmental change. Science 292, 281–284. doi: 10.1126/science.1057544
Trivedi, P., Delgado-Baquerizo, M., Trivedi, C., Hu, H., Anderson, I. C., Jeffries, T. C., et al. (2016). Microbial regulation of the soil carbon cycle: evidence from gene–enzyme relationships. ISME J. 10, 2593–2604. doi: 10.1038/ismej.2016.65
Tschumi, M., Albrecht, M., Bärtschi, C., Collatz, J., Entling, M. H., and Jacot, K. (2016). Perennial, species-rich wildflower strips enhance pest control and crop yield. Agric. Ecosyst. Environ. 220, 97–103. doi: 10.1016/j.agee.2016.01.001
van Bniggen, A. H. C., and Termorskuizen, A. J. (2003). Integrated approaches to root disease management in organic farming systems. Australas. Plant Pathol. 32, 141–156. doi: 10.1071/AP03029
van der Heijden, M. G. A., Bardgett, R. D., and van Straalen, N. M. (2008). The unseen majority: soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310. doi: 10.1111/j.1461-0248.2007.01139.x
Verbruggen, E., Röling, W. F. M., Gamper, H. A, Kowalchuk, G. A, Verhoef, H. A, and van der Heijden, M. G. A (2010). Positive effects of organic farming on below-ground mutualists: large-scale comparison of mycorrhizal fungal communities in agricultural soils. N. Phytol. 186, 968–979. doi: 10.1111/j.1469-8137.2010.03230.x
Wezel, A., Bellon, S., Doré, T., Francis, C., Vallod, D., and David, C. (2009). Agroecology as a science, a movement and a practice. A review. Agron. Sustain. Dev. 29, 503–515. doi: 10.1051/agro/2009004
Wezel, A., Casagrande, M., Celette, F., Vian, J.-F., Ferrer, A., and Peigné, J. (2014). Agroecological practices for sustainable agriculture. A review. Agron. Sustain. Dev. 34, 1–20. doi: 10.1007/s13593-013-0180-7
Wrighton, K. C., Thomas, B. C., Sharon, I., Miller, C. S., Castelle, C. J., VerBerkmoes, N. C., et al. (2012). Fermentation,hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337, 1661–1665. doi: 10.1126/science.1224041
Keywords: sustainable agriculture, soil microbial diversity, bacteria, fungi, metabarcoding, 16S rRNA genes, internal transcribed spacers, soil texture
Citation: Degrune F, Boeraeve F, Dufrêne M, Cornélis J-T, Frey B and Hartmann M (2019) The Pedological Context Modulates the Response of Soil Microbial Communities to Agroecological Management. Front. Ecol. Evol. 7:261. doi: 10.3389/fevo.2019.00261
Received: 01 April 2019; Accepted: 20 June 2019;
Published: 12 July 2019.
Edited by:
Sudhakar Srivastava, Banaras Hindu University, IndiaReviewed by:
Saurabh Yadav, Hemwati Nandan Bahuguna Garhwal University, IndiaRobert Walter Buchkowski, Yale University, United States
Copyright © 2019 Degrune, Boeraeve, Dufrêne, Cornélis, Frey and Hartmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Martin Hartmann, martin.hartmann@usys.ethz.ch
 Florine Degrune
Florine Degrune Fanny Boeraeve
Fanny Boeraeve Marc Dufrêne
Marc Dufrêne Jean-Thomas Cornélis
Jean-Thomas Cornélis Beat Frey
Beat Frey Martin Hartmann
Martin Hartmann