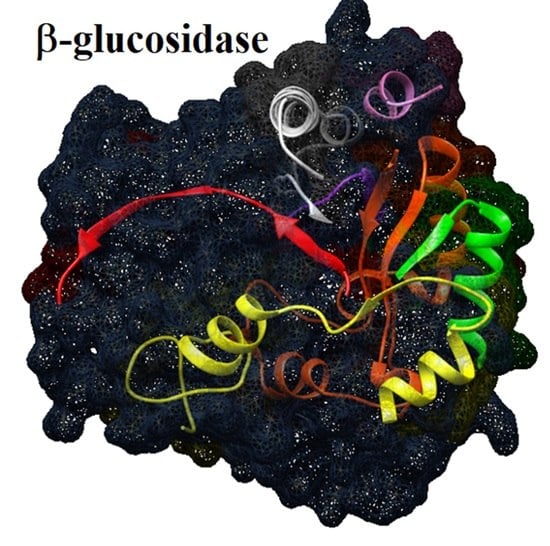
Aggregation and Molecular Properties of β-Glucosidase Isoform II in Chayote (Sechium edule)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation of Crude Extract from Sechium edule
2.2. Extraction and Purification of β-Glucosidase II (Chayote Pulp Homogenate)
2.3. Purification of β-Glucosidase (Sechium edule) through Cation Exchange Chromatography
2.4. Purification of β-Glucosidase (Sechium edule) by Gel Filtration
2.5. Purification of Sechium edule β-Glucosidase by Anion Exchange Chromatography and Detection of Molecular Aggregates
2.6. SDS-PAGE Electrophoresis
2.7. Native-PAGE Electrophoresis
2.8. Electrophoretic Analysis in Acetate-Urea pH 4.4
2.9. Analysis in Two-Dimensional Electrophoresis
2.10. Specificity Assays with Artificial Substrates and Kinetic Parameters
2.11. Analysis of Tryptic Peptides
2.12. Molecular Aggregates
3. Experimental
3.1. Preparation of the Modified Crude Extract of Sechium Edule
3.2. Enzyme Purification
3.2.1. Cation Exchange Chromatography
3.2.2. Molecular Exclusion Chromatography
3.2.3. Anionic Exchange Chromatography
3.2.4. Dialysis and Lyophilization
3.3. Determination of Molecular Mass and Identification of Molecular Aggregates in SDS-PAGE
3.3.1. Native-PAGE Electrophoresis
3.3.2. Acetate-Urea Acid Electrophoresis
3.3.3. Two-Dimensional Electrophoresis
3.4. Determination of Partial Amino Acid Sequence Using Nano-LC-ESI-MS/MS
3.5. Bioinformatic Analysis
3.6. Enzymatic Activity
3.7. Determination of Kinetic Parameters
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Walz, C.; Giavalisco, P.; Schad, M.; Juenger, M.; Klose, J.; Kehr, J. Proteomics of curcurbit phloem exudate reveals a network of defence proteins. Phytochemistry 2004, 65, 1795–1804. [Google Scholar] [CrossRef] [PubMed]
- Malboobi, M.A.; Lefebvre, D.D. A phosphate-starvation inducible beta-glucosidase gene (psr3.2) isolated from Arabidopsis thaliana is a member of a distinct subfamily of the BGA family. Plant Mol. Biol. 1997, 34, 57–68. [Google Scholar] [CrossRef] [PubMed]
- van de Ven, W.T.; LeVesque, C.S.; Perring, T.M.; Walling, L.L. Local and systemic changes in squash gene expression in response to silver winged whitefly feeding. Plant Cell 2000, 12, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, S.; Borchert, C.; Deyholos, M.; Wang, H.; Brazille, S.; Kawai, K.; Galbraith, D.; Bohnert, H.J. Gene expression profiles during the initial phase of salt stress in rice. Plant Cell 2001, 13, 889–905. [Google Scholar] [CrossRef] [Green Version]
- Thorlby, G.; Fourier, N.; Warren, G. The SENSITIVE TO FREEZING2 gene, required for freezing tolerance in Arabidopsis thaliana, encodes a β-glucosidase. Plant Cell 2004, 16, 2192–2203. [Google Scholar] [CrossRef] [Green Version]
- Lipka, V.; Dittgen, J.; Bednarek, P.; Bhat, R.; Wiermer, M.; Stein, M.; Landtag, J.; Brandt, W.; Rosahl, S.; Scheel, D.; et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 2005, 310, 1180–1183. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Li, P.; Sun, L.; Wang, Y.; Ji, K.; Sun, Y.; Leng, P. Expression analysis of β-glucosidase genes that regulate abscisic acid homeostasis during watermelon (Citrullus lanatus) development and under stress conditions. J. Plant Physiol. 2012, 169, 78–85. [Google Scholar] [CrossRef]
- Rachmawati, Y.; Aristya, G.R.; Daryono, B.S. CmBGI Gene Expression encoding β-glucosidase in melon (Cucumis melo L.) under stress condition. Biotropic J. Trop. Biol. 2017, 1, 1–8. [Google Scholar] [CrossRef]
- Hamissou, M.; Smith, A.C.; Carter Jr, R.E.; Triplett II, J.K. Antioxidative properties of bitter gourd (Momordica charantia) and zucchini (Cucurbita pepo). Emir. J. Food Agric. 2013, 25, 641–647. [Google Scholar] [CrossRef] [Green Version]
- Espíndola-Mateos, S.; Cervantes, C.A.; Zenteno, E.; Slomianny, M.C.; Alpuche, J.; Hernández-Cruz, P.; Martínez-Cruz, R.; Pina-Canseco, M.S.; Pérez-Campos, E.; Rubio, M.S.; et al. Purification and partial characterization of β-Glucosidase in Chayote (Sechium edule). Molecules 2015, 20, 19372–19392. [Google Scholar] [CrossRef] [Green Version]
- Poungbangpho, S.; Chuasuwan, S.; Chuphayak, C. Screening of β-D-galactosidase, β-D-glucosidase, and alpha-D-mannosidase in the seeds of cucurbitaceae. Naresuan Univ. 2000, 8, 56–67. [Google Scholar]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280, 309–316. [Google Scholar] [CrossRef] [PubMed]
- Henrissat, B.; Davies, G. Structural and sequence-based classification of glycosyl hydrolases. Curr. Opin. Struct. Biol. 1997, 7, 637–644. [Google Scholar] [CrossRef]
- Rosales-Calderon, O.; Trajano, H.L.; Duff, S.J. Stability of commercial glucanase and β-glucosidase preparations under hydrolysis conditions. PeerJ 2014, 2, e402. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.S.D.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef] [Green Version]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodriguez, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization–purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef] [Green Version]
- Castro-Rodríguez, J.M.; Toledo-Díaz, A.M.; Rodríguez-Galdón, B.; Perdomo-Molina, A.; Rodríguez-Rodríguez, E.M.; Díaz-Romero, C. Caracterización morfológica y composición química de chayotas (Sechium edule) cultivadas en las Islas Canarias (España). Arch. Latinoam. De Nutr. 2015, 65, 243–253. Available online: http://www.alanrevista.org/ediciones/2015/4/art-5/ (accessed on 15 November 2019).
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. Prelude: Biochemistry and the Genomic Revolution. In Biochemistry, 5th ed.; W.H. Freeman and Company: New York, NY, USA, 2002; pp. 14–15, ISBN-10: 0-7167-3051-0. [Google Scholar]
- Vassão, D.G.; Wielsch, N.; Gomes, A.M.D.M.M.; Gebauer-Jung, S.; Hupfer, Y.; Svatoš, A.; Gershenzon, J. Plant defensive β-glucosidases resist digestion and sustain activity in the gut of a lepidopteran herbivore. Front. Plant Sci. 2018, 9, 1389. [Google Scholar] [CrossRef]
- Yamada, K.; Hara-Nishimura, I.; Nishimura, M. Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol. 2011, 52, 2039–2049. [Google Scholar] [CrossRef] [Green Version]
- Ketudat-Cairns, J.R.; Esen, A. β-Glucosidases. Cell Mol. Life Sci. 2010, 67, 3389–3405. [Google Scholar] [CrossRef]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cicek, M.; Esen, A. Expression of soluble and catalytically active plant (monocot) β-glucosidases in E. coli. Biotechnol. Bioeng. 1999, 63, 392–400. [Google Scholar] [CrossRef]
- Cicek, M.; Esen, A. Structure and expression of a dhurrinase (β-glucosidase) from sorghum. Plant Physiol. 1998, 116, 1469–1478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gus-Mayer, S.; Brunner, H.; Schneider-Poetsch, H.A.; Rüdiger, W. Avenacosidase from oat: Purification, sequence analysis and biochemical characterization of a new member of the BGA family of β-glucosidases. Plant Mol. Biol. 1994, 26, 909–921. [Google Scholar] [CrossRef] [PubMed]
- Opassiri, R.; Cairns, J.R.K.; Akiyama, T.; Wara-Aswapati, O.; Svasti, J.; Esen, A. Characterization of a rice β-glucosidase highly expressed in flower and germinating shoot. Plant Sci. 2003, 165, 627–638. [Google Scholar] [CrossRef]
- Kittur, F.S.; Lalgondar, M.; Yu, H.Y.; Bevan, D.R.; Esen, A. Maize β-glucosidase-aggregating factor is a polyspecific jacalin-related chimeric lectin, and its lectin domain is responsible for β-glucosidase aggregation. J. Biol. Chem. 2007, 282, 7299–7311. [Google Scholar] [CrossRef] [Green Version]
- Molina, J.; Landa, A.; Bautista, G.; Martínez, M.; Córdoba, F. Molecular association of lectin and β-glucosidase in corn coleoptile. Biochim. Et Biophys. Acta (Bba)-Gen. Subj. 2004, 1674, 299–304. [Google Scholar] [CrossRef]
- Vozari-Hampe, M.M.; Viegas, C.; Saucedo, C.; Rosseto, S.; Manica, G.G.; Hampe, O.G. A lectin from Sechium edule fruit exudate. Phytochemistry 1992, 31, 1477–1480. [Google Scholar] [CrossRef]
- González-Lucas, J.; Báez-Santacruz, J.; Serna-Lagunes, R.; Llarena-Hernández, R.C.; Núñez-Pastrana, R.; Reynoso-Velasco, D. Phytophagous true bugs (Hemiptera: Heteroptera) associate with chayote crops (Sechium edule jacq) in central Veracruz México. Entomol. Agrícola 2019, 6, 170–176. [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for quantitation of micrograms quantities of proteins utilizing the principles of protein dye-binding. Anal. Biochemistry 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Esen, A. Purification and partial characterization of maize (Zea mays L.) β-glucosidase. Plant Physiol. 1992, 98, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophase T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Hedrick, J.L.; Smith, A.J. Size and charge isomer separation and estimation of molecular weights of proteins by disc gel electrophoresis. Arch. Biochem. Biophys. 1968, 126, 155–164. [Google Scholar] [CrossRef]
- Ferguson, K.A. Starch-gel electrophoresis-application to the classification of pituitary proteins and polypeptides. Metabolism 1964, 13, 985–1002. [Google Scholar] [CrossRef]
- Reisfeld, R.A.; Lewis, U.; Williams, D.E. Disk electrophoresis of basic proteins and peptides on polyacrylamide gels. Nature 1962, 195, 281–283. [Google Scholar] [CrossRef] [PubMed]
- O’Farrell, P.H. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 1975, 250, 4007–4021. [Google Scholar]
- Findlay, J.B.; Pappin, D.J.; Keen, J. Automated solid-phase microsequencing. In Protein Sequencing: A Practical Approach; Findlay, J.B., Geisow, M.J., Eds.; IRL Publishers: Oxford, UK, 1989; pp. 69–84. ISBN1 0199630135. ISBN2 9780199630134. [Google Scholar]
- Zhang, D.M.; Feng, L.X.; Li, L.; Liu, M.; Jiang, B.H.; Yang, M.; Li, G.Q.; Wu, W.Y.; Guo, D.A.; Liu, X. Nano-LC-ESI MS/MS analysis of proteins in dried sea dragon Solenognathus hardwickii and bioinformatic analysis of its protein expression profiling. Chin. J. Nat. Med. 2016, 14, 709–713. [Google Scholar] [CrossRef]
Sample Availability: Samples of the β-Glucosidase from chayote (Sechium edule) are available from the authors. |











| Fraction | Total Protein (mg) | Activity of β-Glucosidase (U) | Specific Activity (U/mg) | Purification Factor | Yield (%) |
|---|---|---|---|---|---|
| Crude pulp extract | 1565.58 | 12,037.56 | 7.69 | 1 | 100 |
| Glacial acetic acid Protein supernatant | 445.36 | 10,479.30 | 23.53 | 3.05 | 87.05 |
| Cationic chromatography CMC | 19.25 | 4276.20 | 222.14 | 28.88 | 36 |
| Gel filtration (S-200HR) | 5.107 | 4102.20 | 803.25 | 104.45 | 34 |
| Anionic chromatography (QFF) | 4.855 | 4085.90 | 841.58 | 109.36 | 33.90 |
| Amino Acids | Res/mol | Residues/100% | |
|---|---|---|---|
| Val | V | 4 | 2.23 |
| Phe | F | 12 | 6.70 |
| Gly | G | 14 | 7.82 |
| Ser | S | 7 | 3.91 |
| Ala | A | 16 | 8.94 |
| Tyr | Y | 17 | 9.49 |
| Gln | Q | 4 | 2.23 |
| Glu | E | 10 | 5.58 |
| Asp | D | 12 | 6.70 |
| Lys | K | 12 | 6.70 |
| Asn | N | 13 | 7.26 |
| Ile | I | 10 | 5.58 |
| Trp | W | 4 | 2.23 |
| Thr | T | 8 | 4.46 |
| His | H | 8 | 4.46 |
| Pro | P | 7 | 3.91 |
| Arg | R | 4 | 2.23 |
| Leu | L | 14 | 7.82 |
| Met | M | 3 | 1.67 |
| Totals | 179 | 100 | |
| Artificial Chromogenic Specific Substrates |
| p-Nitrophenyl-β-d-glucopyranoside |
| p-Nitrophenyl-β-d-galactopyranoside |
| p-Nitrophenyl-β-d-fucopyranoside |
| Non-Specific Substrates |
| p-Nitrophenyl-β-d-manopyranoside |
| p-Nitrophenyl-β-d-lactopyranoside |
| p-Nitrophenyl-β-d-maltopyranoside |
| p-Nitrophenyl-α-d-glucopyranoside |
| p-Nitrophenyl-β-d-N,N′-diacetylcytotriose |
| p-Nitrophenyl-β-d-cellobioside |
| p-Nitrophenyl-β-d-N-acetylgalactosamine |
| Substrate | Km (mM) | Standard Error Km | kcat (min−1) | Standard Error kcat | 10−3 × kcat/Km (mM−1 min−1) |
|---|---|---|---|---|---|
| pNPGlc | 4.59 | 0.49 | 10,086 | 1008 | 2197 |
| pNPGal | 5.72 | 0.45 | 13,718 | 1070 | 2398 |
| pNPFuc | 16.11 | 2.62 | 16,289 | 2638 | 1011 |
| Name | Ion | m/z1 | MW 2 (Da) | Sequence | Start-End Sequence % Identical |
|---|---|---|---|---|---|
| Sebg1 | 1129.01 | 2 | 2256.02 | VFGSASAAYQFEGAAFEDGK | 46-65-Cmbg 100% y Csbg 85% |
| Sebg2 | 903.42 | 3 | 2702.26 | NIWDTFTHKHPTR | 68-80-Cmbg 62% y Csbg 54% |
| Sebg3 | 942.53 | 2 | 1883.06 | IYDHSDGDVALDQYHR | 81-96-Cmbg 94% y Csbg 81.25% |
| Sebg4 | 408.89 | 3 | 1223.67 | YKEDVALMKK | 97-106-Cmbg 90% y Csbg 80% |
| Sebg5 | 122.63 | 2 | 2439.26 | EYYNNLINELLANGIQP | 137-153-Cmbg 94% y Csbg 88.23% |
| Sebg6 | 1391.63 | 2 | 2781.26 | HWITFNEPWSFSMGGYAQGANAPGR | 199-223-Cmbg 96% y Csbg 88% |
| Sebg7 | 538.82 | 2 | 1075.64 | SLPKFSAK | 324-332-Cmbg 75% y Csbg 50% |
| Sebg8 | 1164.07 | 2 | 2326.14 | NALDFLGLNYYTANYAK | 338-354-Cmbg 88% y Csbg 76% |
| Sebg9 | 1333.14 | 2 | 2664.28 | IYITENGYLEIDGPPFHEMGIADK | 416-439-Cmbg 62.5% y Csbg 62.5% |
| Sebg10 | 811.92 | 2 | 1621.84 | KV-YYHDHLYNLR | 440-451- Cmbg 66.6% y Csbg 50% |
| Sebg11 | 560.29 | 2 | 1118.58 | FGLTYIDYK | 483-491-Cmbg 100% y Csbg100% |
| Sebg12 | 542.57 | 2 | 982.49 | WFENFLKT | 504-511-Cmbg 100% y Csbg 87.5% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cruz Rodríguez, A.; Sánchez Esperanza, F.A.; Pérez-Campos, E.; Hernández-Huerta, M.T.; Pérez-Campos Mayoral, L.; Matias-Cervantes, C.A.; Martínez Barras, A.; Mayoral-Andrade, G.; Santos Pineda, L.Á.; Díaz Barrita, A.J.; et al. Aggregation and Molecular Properties of β-Glucosidase Isoform II in Chayote (Sechium edule). Molecules 2020, 25, 1699. https://doi.org/10.3390/molecules25071699
Cruz Rodríguez A, Sánchez Esperanza FA, Pérez-Campos E, Hernández-Huerta MT, Pérez-Campos Mayoral L, Matias-Cervantes CA, Martínez Barras A, Mayoral-Andrade G, Santos Pineda LÁ, Díaz Barrita AJ, et al. Aggregation and Molecular Properties of β-Glucosidase Isoform II in Chayote (Sechium edule). Molecules. 2020; 25(7):1699. https://doi.org/10.3390/molecules25071699
Chicago/Turabian StyleCruz Rodríguez, Alberto, Fabiola Anaid Sánchez Esperanza, Eduardo Pérez-Campos, María Teresa Hernández-Huerta, Laura Pérez-Campos Mayoral, Carlos Alberto Matias-Cervantes, Alexis Martínez Barras, Gabriel Mayoral-Andrade, Luis Ángel Santos Pineda, Aymara Judith Díaz Barrita, and et al. 2020. "Aggregation and Molecular Properties of β-Glucosidase Isoform II in Chayote (Sechium edule)" Molecules 25, no. 7: 1699. https://doi.org/10.3390/molecules25071699