Extracellular Vesicles in Human Preterm Colostrum Inhibit Infection by Human Cytomegalovirus In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Colostrum Collection
2.2. Colostrum Clarification and Extracellular Vesicle (EV) Isolation
2.3. EV Characterization by Immunoblotting
2.4. Nanoparticle Tracking Analysis (NTA)
2.5. Cell lines and Viruses
2.6. Cell Viability Assay
2.7. HCMV Inhibition Assay
2.8. HCMV Inactivation Assay
2.9. Pre-Treatment Assay
2.10. Attachment Assay
2.11. Entry Assay
2.12. Post-Entry Assay
2.13. Data Analysis
2.14. Shaving of Extracellular Vesicles
2.15. ESI-Q-TOF Analysis of Peptides
2.16. Peptide Data Search
2.17. In Silico Analysis of Proteins
3. Results and Discussion
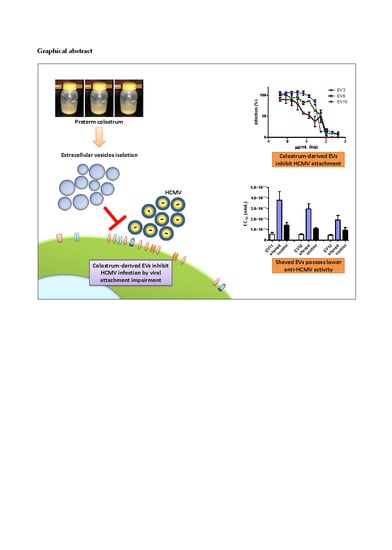
3.1. Milk Sample Collection and Antiviral Activity of Colostrum-Derived Extracellular Vesicles (EVs) against HCMV
3.2. Colostrum-Derived Extracellular Vesicles Inhibit the Attachment of HCMV on Cells
3.3. Shaving of Extracellular Vesicles Significantly Reduced Their Anti-HCMV Activity
3.4. Proteomic Analysis of EV Surfaceome
3.5. Surface Peptide Mixtures Inhibit HCMV Attachment
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO | Guidelines on Optimal Feeding of Low Birth-Weight Infants in Low- and Middle-Income Countries. Available online: http://www.who.int/maternal_child_adolescent/documents/infant_feeding_low_bw/en/ (accessed on 13 March 2020).
- Civardi, E.; Tzialla, C.; Baldanti, F.; Strocchio, L.; Manzoni, P.; Stronati, M. Viral Outbreaks in Neonatal Intensive Care Units: What We Do Not Know. Am. J. Infect. Control 2013, 41, 854–856. [Google Scholar] [CrossRef] [PubMed]
- Eidelman, A.I. Breastfeeding and the Use of Human Milk: An Analysis of the American Academy of Pediatrics 2012 Breastfeeding Policy Statement. Breastfeed. Med. 2012, 7, 323–324. [Google Scholar] [CrossRef] [PubMed]
- Moro, G.E.; Billeaud, C.; Rachel, B.; Calvo, J.; Cavallarin, L.; Christen, L.; Escuder-Vieco, D.; Gaya, A.; Lembo, D.; Wesolowska, A.; et al. Processing of Donor Human Milk: Update and Recommendations From the European Milk Bank Association (EMBA). Front. Pediatr. 2019, 7, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamprecht, K.; Goelz, R. Postnatal Cytomegalovirus Infection Through Human Milk in Preterm Infants: Transmission, Clinical Presentation, and Prevention. Clin. Perinatol. 2017, 44, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Manicklal, S.; Emery, V.C.; Lazzarotto, T.; Boppana, S.B.; Gupta, R.K. The “Silent” Global Burden of Congenital Cytomegalovirus. Clin. Microbiol. Rev. 2013, 26, 86–102. [Google Scholar] [CrossRef] [Green Version]
- Doctor, S.; Friedman, S.; Dunn, M.S.; Asztalos, E.V.; Wylie, L.; Mazzulli, T.; Vearncombe, M.; O’Brien, K. Cytomegalovirus Transmission to Extremely Low-Birthweight Infants through Breast Milk. Acta Paediatr. 2005, 94, 53–58. [Google Scholar] [CrossRef]
- Lanzieri, T.M.; Dollard, S.C.; Josephson, C.D.; Schmid, D.S.; Bialek, S.R. Breast Milk-Acquired Cytomegalovirus Infection and Disease in VLBW and Premature Infants. Pediatrics 2013, 131, e1937–e1945. [Google Scholar] [CrossRef] [Green Version]
- Donalisio, M.; Rittà, M.; Tonetto, P.; Civra, A.; Coscia, A.; Giribaldi, M.; Cavallarin, L.; Moro, G.E.; Bertino, E.; Lembo, D. Anti-Cytomegalovirus Activity in Human Milk and Colostrum From Mothers of Preterm Infants. J. Pediatr. Gastroenterol. Nutr. 2018, 67, 654–659. [Google Scholar] [CrossRef]
- Trend, S.; Strunk, T.; Lloyd, M.L.; Kok, C.H.; Metcalfe, J.; Geddes, D.T.; Lai, C.T.; Richmond, P.; Doherty, D.A.; Simmer, K.; et al. Levels of Innate Immune Factors in Preterm and Term Mothers’ Breast Milk during the 1st Month Postpartum. Br. J. Nutr. 2016, 115, 1178–1193. [Google Scholar] [CrossRef] [Green Version]
- Clarke, N.M.; May, J.T. Effect of Antimicrobial Factors in Human Milk on Rhinoviruses and Milk-Borne Cytomegalovirus In Vitro. J. Med. Microbiol. 2000, 49, 719–723. [Google Scholar] [CrossRef] [Green Version]
- Ng, T.B.; Cheung, R.C.F.; Wong, J.H.; Wang, Y.; Ip, D.T.M.; Wan, D.C.C.; Xia, J. Antiviral Activities of Whey Proteins. Appl. Microbiol. Biotechnol. 2015, 99, 6997–7008. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, H.; Oda, H.; Yamauchi, K.; Abe, F. Lactoferrin for Prevention of Common Viral Infections. J. Infect. Chemother. 2014, 20, 666–671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zempleni, J.; Aguilar-Lozano, A.; Sadri, M.; Sukreet, S.; Manca, S.; Wu, D.; Zhou, F.; Mutai, E. Biological Activities of Extracellular Vesicles and Their Cargos from Bovine and Human Milk in Humans and Implications for Infants. J. Nutr. 2017, 147, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, Biogenesis and Function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Xanthou, M. Immune Protection of Human Milk. Biol. Neonate 1998, 74, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with Immune Modulatory Features Are Present in Human Breast Milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Hock, A.; Miyake, H.; Li, B.; Lee, C.; Ermini, L.; Koike, Y.; Chen, Y.; Määttänen, P.; Zani, A.; Pierro, A. Breast Milk-Derived Exosomes Promote Intestinal Epithelial Cell Growth. J. Pediatr. Surg. 2017, 52, 755–759. [Google Scholar] [CrossRef]
- Näslund, T.I.; Paquin-Proulx, D.; Paredes, P.T.; Vallhov, H.; Sandberg, J.K.; Gabrielsson, S. Exosomes from Breast Milk Inhibit HIV-1 Infection of Dendritic Cells and Subsequent Viral Transfer to CD4+ T Cells. AIDS 2014, 28, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Arslanoglu, S.; Bertino, E.; Tonetto, P.; De Nisi, G.; Ambruzzi, A.M.; Biasini, A.; Profeti, C.; Spreghini, M.R.; Moro, G.E.; Italian Association of Human Milk Banks Associazione Italiana Banche del Latte Umano Donato (AIBLUD: www.aiblud.org). Guidelines for the Establishment and Operation of a Donor Human Milk Bank. J. Matern. Fetal Neonatal Med. 2010, 23, 1–20. [Google Scholar] [CrossRef]
- Marchini, A.; Liu, H.; Zhu, H. Human Cytomegalovirus with IE-2 (UL122) Deleted Fails to Express Early Lytic Genes. J. Virol. 2001, 75, 1870–1878. [Google Scholar] [CrossRef] [Green Version]
- Cagno, V.; Donalisio, M.; Civra, A.; Cagliero, C.; Rubiolo, P.; Lembo, D. In Vitro Evaluation of the Antiviral Properties of Shilajit and Investigation of Its Mechanisms of Action. J. Ethnopharmacol. 2015, 166, 129–134. [Google Scholar] [CrossRef]
- Qin, W.; Tsukasaki, Y.; Dasgupta, S.; Mukhopadhyay, N.; Ikebe, M.; Sauter, E.R. Exosomes in Human Breast Milk Promote EMT. Clin. Cancer Res. 2016, 22, 4517–4524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Q.; Li, M.; Wang, X.; Li, Q.; Wang, T.; Zhu, Q.; Zhou, X.; Wang, X.; Gao, X.; Li, X. Immune-Related MicroRNAs Are Abundant in Breast Milk Exosomes. Int. J. Biol. Sci. 2012, 8, 118–123. [Google Scholar] [CrossRef] [PubMed]
- ExoCarta: Exosome Markers. Available online: http://exocarta.org/exosome_markers (accessed on 13 March 2020).
- Simons, M.; Raposo, G. Exosomes-Vesicular Carriers for Intercellular Communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Welch, J.L.; Kaddour, H.; Winchester, L.; Fletcher, C.V.; Stapleton, J.T.; Okeoma, C.M. Semen Extracellular Vesicles From HIV-1-Infected Individuals Inhibit HIV-1 Replication In Vitro, and Extracellular Vesicles Carry Antiretroviral Drugs In Vivo. J. Acquir. Immune Defic. Syndr. 2020, 83, 90–98. [Google Scholar] [CrossRef]
- Kesimer, M.; Scull, M.; Brighton, B.; DeMaria, G.; Burns, K.; O’Neal, W.; Pickles, R.J.; Sheehan, J.K. Characterization of Exosome-like Vesicles Released from Human Tracheobronchial Ciliated Epithelium: A Possible Role in Innate Defense. FASEB J. 2009, 23, 1858–1868. [Google Scholar] [CrossRef] [Green Version]
- Nolte-’t Hoen, E.; Cremer, T.; Gallo, R.C.; Margolis, L.B. Extracellular Vesicles and Viruses: Are They Close Relatives? Proc. Natl. Acad. Sci. USA 2016, 113, 9155–9161. [Google Scholar] [CrossRef] [Green Version]
- Van Herwijnen, M.J.C.; Zonneveld, M.I.; Goerdayal, S.; Hoen, E.N.M.N.; Garssen, J.; Stahl, B.; Maarten Altelaar, A.F.; Redegeld, F.A.; Wauben, M.H.M. Comprehensive proteomic analysis of human milk-derived extracellular vesicles unveils a novel functional proteome distinct from other milk components. Mol. Cell. Proteom. 2016, 15, 3412–3423. [Google Scholar] [CrossRef] [Green Version]
- Dudkina, N.V.; Spicer, B.A.; Reboul, C.F.; Conroy, P.J.; Lukoyanova, N.; Elmlund, H.; Law, R.H.P.; Ekkel, S.M.; Kondos, S.C.; Goode, R.J.A.; et al. Structure of the poly-C9 component of the complement membrane attack complex. Nat. Commun. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- Schiela, B.; Bernklau, S.; Malekshahi, Z.; Deutschmann, D.; Koske, I.; Banki, Z.; Thielens, N.M.; Würzner, R.; Speth, C.; Weiss, G.; et al. Active human complement reduces the zika virus load via formation of the membrane-attack complex. Front. Immunol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, M.; Muranishi, H.; Yamada, K.; Kakehi, K.; Uchida, K.; Suzuki, T.; Yabe, T.; Nakagomi, T.; Nakagomi, O.; Kanamaru, Y. Bovine κ-casein inhibits human rotavirus (HRV) infection via direct binding of glycans to HRV. J. Dairy Sci. 2014, 97, 2653–2661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mistry, N.; Drobni, P.; Näslund, J.; Sunkari, V.G.; Jenssen, H.; Evander, M. The anti-papillomavirus activity of human and bovine lactoferricin. Antivir. Res. 2007, 75, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Swart, P.J.; Kuipers, E.M.; Smit, C.; Van Der Strate, B.W.A.; Harmsen, M.C.; Meijer, D.K.F. Lactoferrin. In Advances in Lactoferrin Research; Spik, G., Legrand, D., Mazurier, J., Pierce, A., Perraudin, J., Eds.; Advances in Experimental Medicine and Biology, vol 443; Springer: Boston, MA, USA, 1998. [Google Scholar]
- Andersen, J.H.; Osbakk, S.A.; Vorland, L.H.; Traavik, T.; Gutteberg, T.J. Lactoferrin and cyclic lactoferricin inhibit the entry of human cytomegalovirus into human fibroblasts. Antivir. Res. 2001, 51, 141–149. [Google Scholar] [CrossRef]
- Iskarpatyoti, J.A.; Morse, E.A.; Paul McClung, R.; Ikizler, M.; Wetzel, J.D.; Contractor, N.; Dermody, T.S. Serotype-specific differences in inhibition of reovirus infectivity by human-milk glycans are determined by viral attachment protein σ1. Virology 2012, 433, 489–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeland, E.; de Jong, M.A.W.P.; Nabatov, A.A.; Kalay, H.; Geijtenbeek, T.B.H.; van Kooyk, Y. MUC1 in human milk blocks transmission of human immunodeficiency virus from dendritic cells to T cells. Mol. Immunol. 2009, 46, 2309–2316. [Google Scholar] [CrossRef] [PubMed]
- Mall, A.S.; Habte, H.; Mthembu, Y.; Peacocke, J.; De Beer, C. Mucus and Mucins: Do they have a role in the inhibition of the human immunodeficiency virus? Virol. J. 2017, 14, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Dang, L.; Li, D.; Qi, J.; Wang, M.; Chai, W.; Zhang, Q.; Wang, H.; Bai, R.; Tan, M.; et al. Structural Basis of Glycan Recognition in Globally Predominant Human P[8] Rotavirus. Virol. Sin. 2020, 35, 156–170. [Google Scholar] [CrossRef]
- Peterson, J.A.; Patton, S.; Hamosh, M. Glycoproteins of the Human Milk Fat Globule in the Protection of the Breast-Fed Infant against Infections. Neonatology 1998, 74, 143–162. [Google Scholar] [CrossRef]
- Newburg, D.S. Neonatal protection by an innate immune system of human milk consisting of oligosaccharides and glycans. J. Anim. Sci. 2009, 87, 26–34. [Google Scholar] [CrossRef]
- Wyatt, A.; Yerbury, J.; Poon, S.; Dabbs, R.; Wilson, M. Chapter 6: The Chaperone Action of Clusterin and Its Putative Role in Quality Control of Extracellular Protein Folding. Adv. Cancer Res. 2009, 104, 89–114. [Google Scholar] [CrossRef]
- Sabatte, J.; Faigle, W.; Ceballos, A.; Morelle, W.; Rodríguez Rodrígues, C.; Remes Lenicov, F.; Thépaut, M.; Fieschi, F.; Malchiodi, E.; Fernández, M.; et al. Semen Clusterin Is a Novel DC-SIGN Ligand. J. Immunol. 2011, 187, 5299–5309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Švajger, U.; Anderluh, M.; Jeras, M.; Obermajer, N. C-type lectin DC-SIGN: An adhesion, signalling and antigen-uptake molecule that guides dendritic cells in immunity. Cell. Signal. 2010, 22, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Batra, J.; Cao, W.; Sharma, K.; Patel, J.R.; Ranjan, P.; Kumar, A.; Katz, J.M.; Cox, N.J.; Lal, R.B.; et al. Influenza A Virus Nucleoprotein Induces Apoptosis in Human Airway Epithelial Cells: Implications of a Novel Interaction between Nucleoprotein and Host Protein Clusterin. Cell Death Dis. 2013, 4, e562. [Google Scholar] [CrossRef] [Green Version]
- Kurosu, T.; Chaichana, P.; Yamate, M.; Anantapreecha, S.; Ikuta, K. Secreted complement regulatory protein clusterin interacts with dengue virus nonstructural protein 1. Biochem. Biophys. Res. Commun. 2007, 362, 1051–1056. [Google Scholar] [CrossRef] [PubMed]
- Liao, F.T.; Lee, Y.J.; Ko, J.L.; Tsai, C.C.; Tseng, C.J.; Sheu, G.T. Hepatitis delta virus epigenetically enhances clusterin expression via histone acetylation in human hepatocellular carcinoma cells. J. Gen. Virol. 2009, 90, 1124–1134. [Google Scholar] [CrossRef] [PubMed]
- Naarding, M.A.; Dirac, A.M.; Ludwig, I.S.; Speijer, D.; Lindquist, S.; Vestman, E.-L.; Stax, M.J.; Geijtenbeek, T.B.H.; Pollakis, G.; Hernell, O.; et al. Bile Salt-Stimulated Lipase from Human Milk Binds DC-SIGN and Inhibits Human Immunodeficiency Virus Type 1 Transfer to CD4+ T Cells. Antimicrob. Agents Chemother. 2006, 50, 3367–3374. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Newburg, D.S. Human milk glycoproteins protect infants against human pathogens. Breastfeed. Med. 2013, 8, 354–362. [Google Scholar] [CrossRef] [Green Version]
- Azad, A.K.; Rajaram, M.V.S.; Schlesinger, L.S. Exploitation of the Macrophage Mannose Receptor (CD206) in Infectious Disease Diagnostics and Therapeutics. J. Cytol. Mol. Biol. 2014, 1, 1–10. [Google Scholar] [CrossRef]
- Taylor, P.R.; Martinez-Pomares, L.; Stacey, M.; Lin, H.-H.; Brown, G.D.; Gordon, S. Macrophage Receptors and Immune Recognition. Annu. Rev. Immunol. 2005, 23, 901–944. [Google Scholar] [CrossRef]
- Sukegawa, S.; Miyagi, E.; Bouamr, F.; Farkašová, H.; Strebel, K. Mannose Receptor 1 Restricts HIV Particle Release from Infected Macrophages. Cell Rep. 2018, 22, 786–795. [Google Scholar] [CrossRef] [Green Version]
- Cinatl, J., Jr.; Bittoova, M.; Margraf, S.; Vogel, J.; Cinatl, J.; Preiser, W.; Doerr, H.W. Cytomegalovirus Infection Decreases Expression of Thrombospondin-1 and -2 in Cultured Human Retinal Glial Cells: Effects of Antiviral Agents. J. Infect. Dis. 2000, 182, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, Z.; Deng, B.; Wu, X.; Liu, J.; Feng, X. Identification of Enolase 1 and Thrombospondin-1 as serum biomarkers in HBV hepatic fibrosis by proteomics. Proteome Sci. 2013, 11, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, C.C.; Kamil, J.P. Pathogen at the gates: Human cytomegalovirus entry and cell tropism. Viruses 2018, 10, 704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Midwood, K.S.; Hussenet, T.; Langlois, B.; Orend, G. Advances in tenascin-C biology. Cell. Mol. Life Sci. 2011, 68, 3175–3199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouda, G.G.; Jaeger, F.H.; Amos, J.D.; Ho, C.; Kunz, E.L.; Anasti, K.; Stamper, L.W.; Liebl, B.E.; Barbas, K.H.; Ohashi, T.; et al. Tenascin-C Is an Innate Broad-Spectrum, HIV-1-Neutralizing Protein in Breast Milk. Proc. Natl. Acad. Sci. USA 2013, 110, 18220–18225. [Google Scholar] [CrossRef] [Green Version]
- Mangan, R.J.; Stamper, L.; Ohashi, T.; Eudailey, J.A.; Go, E.P.; Jaeger, F.H.; Itell, H.L.; Watts, B.E.; Fouda, G.G.; Erickson, H.P.; et al. Determinants of Tenascin-C and HIV-1 Envelope Binding and Neutralization. Mucosal Immunol. 2019, 12, 1004–1012. [Google Scholar] [CrossRef]
- García-Expósito, L.; Ziglio, S.; Barroso-González, J.; de Armas-Rillo, L.; Valera, M.S.; Zipeto, D.; Machado, J.D.; Valenzuela-Fernández, A. Gelsolin activity controls efficient early HIV-1 infection. Retrovirology 2013, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Zhang, M.; Zhang, L.; Wang, P.; Song, G.; Liu, B.; Wu, H.; Yin, Z.; Gao, C. Intracellular osteopontin stabilizes TRAF3 to positively regulate innate antiviral response. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]








| Sample no. | Parity * | Breastfeeding after Previous Delivery(ies) | Type of Delivery |
|---|---|---|---|
| 1 | 2002 | Yes | CS |
| 2 | 0000 | - | CS |
| 3 | 0000 | - | CS |
| 4 | 1001 | Yes | CS |
| 5 | 0000 | - | S |
| 6 | 1021 | Yes | S |
| 7 | 0000 | - | CS |
| 8 | 0000 | - | CS |
| 9 | 1001 | Yes | CS |
| 10 | 1001 | Yes | CS |
| 11 | 1001 | - | S |
| 12 | 0000 | Yes | CS |
| 13 | 1001 | - | CS |
| Sample no. | EC50a (µg/mL) (95% CI b) | EC90c (µg/mL) (95% CI) | |
|---|---|---|---|
| Colostra | 1 | 67.33 (54.24–83.57) | 190 (118.5–304.5) |
| 2 | 62.58 (50.45–77.63) | 215 (133.9–345.3) | |
| 3d | 153.5 (129.5–182.1) | 527.6 (359.1–775.1) | |
| 4 | 152 (83.46–276.8) | 581.8 (160.1–2115) | |
| 5 | 115.7 (109.3–122.6) | 306 (269.7–347.2) | |
| 6 | 104.3 (84.02–129.5) | 652.9 (404.6–1053) | |
| 7 | 51.35 (28.24–93.38) | 647.4 (170.6–2457) | |
| 8 | 111.8 (90.67–137.9) | 323.2 (204.3–511.6) | |
| 9 | 149.7 (116.9–191.6) | 920.1 (545.3–1552) | |
| 10 | 38.71 (29.82–50.24) | 738.3 (405.4–1345) | |
| 11 | 125.3 (105.9–148.2) | 356.8 (245.1–519.4) | |
| 12 | 140.6 (107.9–198.4) | 637.5 (431.6–941.5) | |
| 13 | 213.9 (186.2–245.7) | 413.9 (290.4–590) | |
| Colostrum-derived EV | 1 | 35.29 (23.91–52.08) | 200.9 (85.08–474.4) |
| 2 | 8.84 (5.66–13.81) | 120.4 (42.95–337.4) | |
| 3d | 41.56 (27.44–62.95) | 222.2 (87.30–565.7) | |
| 4 | 42.16 (27.33–65.02) | 435.9 (168.3–1129) | |
| 5 | 55.55 (49.31–62.58) | 97.31 (69.92–135.4) | |
| 6 | 19.68 (14.09–27.49) | 479.6 (219.7–1047) | |
| 7 | 3.98 (1.36–11.63) | 844.1 (74.99–9501) | |
| 8 | 21.96 (13.20–36.53) | 277.2 (85.72–896.4) | |
| 9 | 75.93 (47.92–120.3) | 625.1 (201.5–1940) | |
| 10 | 33.92 (14.31–80.38) | 1324 (65.48–26763) | |
| 11 | 74.21 (62.6–113.3) | 493.8 (246.8–988) | |
| 12 | 41.4 (32.7–64.59) | 277.6 (159.7–482.4) | |
| 13 | 30.92 (22.27–42.94) | 371 (187.3–735.1) |
| Uniprot Id | Description | MW | pI | Biological Function |
|---|---|---|---|---|
| O00300 | Tumor necrosis factor receptor superfamily member 11B | 45,996 | 8.66 | Signaling pathway |
| O00391 | Sulfhydryl oxidase 1 | 66,818 | 9.13 | Oxidation-reduction process |
| O15232 | Matrilin-3 | 52,816 | 6.25 | Calcium ion binding |
| P00709 | Alpha-lactalbumin | 16,214 | 4.83 | Lactose biosynthetic process |
| P01011 | Alpha-1-antichymotrypsin | 47,621 | 5.33 | Proteolysis |
| P01024 | Complement C3 | 187,030 | 6.02 | Immune response |
| P01833 | Polymeric immunoglobulin receptor | 83,232 | 5.58 | Immune response |
| P01834 | Immunoglobulin kappa constant | 11,758 | 6.11 | Immune response |
| P01876 | Immunoglobulin heavy constant alpha 1 | 37,631 | 5.99 | Immune response |
| P02647 | Apolipoprotein A-I | 30,759 | 5.56 | Lipid transport |
| P02649 | Apolipoprotein E | 36,132 | 5.65 | Lipid transport |
| P02652 | Apolipoprotein A-II | 11,168 | 6.26 | Lipid transport |
| P02748 | Complement component C9 | 63,133 | 5.43 | Immune response |
| P02788 | Lactotransferrin | 78,182 | 8.50 | Immune response |
| P04114 | Apolipoprotein B-100 | 515,283 | 6.58 | Lipid transport |
| P04406 | Glyceraldehyde-3-phosphate dehydrogenase | 36,030 | 8.57 | Glucose metabolic process |
| P05814 | Beta-casein | 25,366 | 5.52 | Casein micelle formation |
| P06396 | Gelsolin | 85,698 | 5.90 | Cell motility and contraction |
| P06858 | Lipoprotein lipase | 53,129 | 8.37 | Lipid metabolic process |
| P07498 | Kappa-casein | 20,293 | 8.97 | Casein micelle formation |
| P07602 | Prosaposin | 58,112 | 5.06 | Lipid metabolic process |
| P07996 | Thrombospondin-1 | 129,300 | 4.71 | Adhesion |
| P08571 | Monocyte differentiation antigen CD14 | 40,051 | 5.84 | Immune response |
| P10451 | Osteopontin | 35,422 | 4.37 | Adhesion |
| P10909 | Clusterin | 52,494 | 5.88 | Protein folding/transport |
| P15291 | Beta-1,4-galactosyltransferase 1 | 43,920 | 8.88 | Carbohydrate metabolic process |
| P19835 | Bile salt-activated lipase | 79,272 | 5.13 | Lipid metabolic process |
| P22897 | Macrophage mannose receptor 1 | 165,905 | 6.11 | Endocytosis |
| P23280 | Carbonic anhydrase 6 | 35,366 | 6.51 | Carbon metabolic process |
| P23284 | Peptidyl-prolyl cis-trans isomerase B | 23,728 | 9.42 | Protein folding/transport |
| P24821 | Tenascin | 240,853 | 4.79 | Adhesion |
| P47710 | Alpha-S1-casein | 21,671 | 5.32 | Calcium ion binding |
| P47989 | Xanthine dehydrogenase/oxidase | 146,330 | 7.86 | Oxidation-reduction process |
| P49327 | Fatty acid synthase | 273,254 | 6.01 | Oxidation-reduction process |
| P58499 | Protein FAM3B | 25,981 | 8.97 | Insulin secretion |
| P60709 | Actin, cytoplasmic 1 | 41,710 | 5.29 | Cell motility and contraction |
| Q02809 | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | 83,550 | 6.46 | Oxidation-reduction process |
| Q08431 | Lactadherin | 43,105 | 8.47 | Adhesion |
| Q13410 | Butyrophilin subfamily 1 member A1 | 58,923 | 5.38 | Milk-fat droplet release / immune response |
| Q14697 | Neutral alpha-glucosidase AB | 106,873 | 5.74 | Carbohydrate metabolic process |
| Q6WN34 | Chordin-like protein 2 | 49,643 | 6.75 | Signaling pathway |
| Q96DA0 | Zymogen granule protein 16 homolog B | 22,725 | 6.74 | Protein folding/transport |
| Q96S86 | Hyaluronan and proteoglycan link protein 3 | 40,868 | 6.07 | Adhesion |
| Q99102 | Isoform 10 of Mucin-4 | 225,293 | 5.85 | Adhesion |
| Q9H173 | Nucleotide exchange factor SIL1 | 52,052 | 5.27 | Protein folding/transport |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donalisio, M.; Cirrincione, S.; Rittà, M.; Lamberti, C.; Civra, A.; Francese, R.; Tonetto, P.; Sottemano, S.; Manfredi, M.; Lorenzato, A.; et al. Extracellular Vesicles in Human Preterm Colostrum Inhibit Infection by Human Cytomegalovirus In Vitro. Microorganisms 2020, 8, 1087. https://doi.org/10.3390/microorganisms8071087
Donalisio M, Cirrincione S, Rittà M, Lamberti C, Civra A, Francese R, Tonetto P, Sottemano S, Manfredi M, Lorenzato A, et al. Extracellular Vesicles in Human Preterm Colostrum Inhibit Infection by Human Cytomegalovirus In Vitro. Microorganisms. 2020; 8(7):1087. https://doi.org/10.3390/microorganisms8071087
Chicago/Turabian StyleDonalisio, Manuela, Simona Cirrincione, Massimo Rittà, Cristina Lamberti, Andrea Civra, Rachele Francese, Paola Tonetto, Stefano Sottemano, Marcello Manfredi, Annalisa Lorenzato, and et al. 2020. "Extracellular Vesicles in Human Preterm Colostrum Inhibit Infection by Human Cytomegalovirus In Vitro" Microorganisms 8, no. 7: 1087. https://doi.org/10.3390/microorganisms8071087