Abstract
The neural crest gives rise to numerous cell types, dysfunction of which contributes to many disorders. Here, we report that adenosine deaminase acting on RNA (ADAR1), responsible for adenosine-to-inosine editing of RNA, is required for regulating the development of two neural crest derivatives: melanocytes and Schwann cells. Neural crest specific conditional deletion of Adar1 in mice leads to global depigmentation and absence of myelin from peripheral nerves, resulting from alterations in melanocyte survival and differentiation of Schwann cells, respectively. Upregulation of interferon stimulated genes precedes these defects, which are associated with the triggering of a signature resembling response to injury in peripheral nerves. Simultaneous extinction of MDA5, a key sensor of unedited RNA, rescues both melanocytes and myelin defects in vitro, suggesting that ADAR1 safeguards neural crest derivatives from aberrant MDA5-mediated interferon production. We thus extend the landscape of ADAR1 function to the fields of neural crest development and disease.
Similar content being viewed by others
Introduction
Adenosine-to-inosine (A-to-I) RNA editing is a well-known contributor of transcriptomic and, to a lesser extent, proteomic diversity1. Because the resulting inosines are subsequently read as guanosines, this post-transcriptional modification can indeed affect protein function, splicing, RNA stability, transcriptional regulation and miRNA processing2,3. A-to-I editing is catalyzed by enzymes of the ADAR family (adenosine deaminase acting on double stranded (ds)RNA; ADAR1-3)4,5. Available data suggest that Adar2 is mainly expressed in brain and is the primary editor of nonrepetitive coding sites6. In contrast, ADAR1 has been shown to be the primary editor of repeat elements (Alu and SINES) in noncoding sequences1.
Adar1 is widely expressed and is the most highly expressed ADAR outside the central nervous system1,4,5,7. In mice, Adar1 deletion is lethal between embryonic days (E)11.5 and 13.5, due to fetal liver disintegration8. Hematopoietic progenitors depend on Adar1 for their survival and maturation8,9. Transcriptional profiling of these cell types has highlighted the activation of interferon (IFN) along with a large number of interferon-stimulated genes (ISGs) upon Adar1 deletion10,11. The embryonic lethality of Adar1 mutants is rescued upon concomitant deletion of either Ifih1 or Mavs, encoding the cytosolic dsRNA sensor MDA5 (melanoma differentiation-associated protein 5) and MAVS (mitochondrial antiviral-signaling protein), respectively10,11,12. Overall, these data suggest that the key in vivo function of ADAR1 is to mark, via editing, endogenous dsRNAs as self and prevent their immune recognition by MDA5, safeguarding cells from unwanted MDA5-mediated IFN production and ISGs expression in a cell-type-specific manner. However, an MDA5-MAVS-independent function for ADAR1 is described in the development of multiple organs11.
In humans, mutations of ADAR1 are responsible for Aicardi-Goutières Syndrome (AGS, OMIM: 61501013,14,15), an inflammatory encephalopathy also referred to as a type 1 interferonopathy, and Dyschromatosis Symmetrica Hereditaria (DSH, OMIM: 12740016,17,18), characterized by hypo- and hyperpigmentation macules on the extremities that appear in infancy. Skin pigmentation defects suggest that ADAR1 is important for melanocytes, at least after birth18,19.
Melanocytes, specialized skin cells that produce the melanin, are one of the numerous cell types that originate from a transient embryonic structure called neural crest (NC)20,21,22. Upon neural tube closure, NC-derived cells migrate throughout the embryo, and differentiate into melanocytes (mostly found in epidermis and hair follicle)20,22, but also into cells contributing to septation of the cardiac outflow tract and ventricles, skeletal and connective tissue components of the head, and most neurons and glia of the peripheral nervous system (PNS) including the enteric nervous system and myelin producing-Schwann cells (SCs)23,24. Along nerves, SCs precursors develop into immature SCs, then into promyelinating SCs, which establish a one-to-one relationship with large caliber axons, before finally transforming into myelin-forming mature SCs within the first two postnatal weeks in mice24,25,26.
The coordinated action of signal transduction cascades, transcription factors and miRNA regulation converge to control the various steps of the development of melanocytes and SCs22,26,27,28,29,30. To explore the involvement of RNA editing in the development of NC cells, we generated mouse lines with NC-cell-specific deletion of Adar1. The mutants show depigmentation and absence of myelin within the peripheral nerves. Presented analyses suggest that ADAR1 safeguards both melanocytes and SCs from unwanted activation of ISGs and subsequent cell survival and differentiation alterations, respectively. Our results thus extend the landscape of the functional roles of this enzyme and A-to-I RNA editing to the fields of NC development and NC-linked disorders.
Results
Depigmentation and early lethality upon NC deletion of Adar1
We studied the requirement of A-to-I RNA editing in NC cells in vivo by breeding mice harboring a floxed (fl) Adar1 allele with mice expressing Cre recombinase driven by the human tissue plasminogen (HtPA) promoter, triggering ablation of the floxed alleles in NC cells from E931. A YFP tracer (R26R) was also incorporated to specifically mark NC cells and their derivatives32 (crosses in Fig. 1a and Methods). At birth (P0), the heterozygous as well as the HtPA-Cre; Adar1fl/fl;R26R mutant animals (referred to as mutants) were present at the expected Mendelian ratio (Fig. 1b) and were not discernable from their wild-type counterparts macroscopically (similar size and weight, Fig. 1c). Four days later (postnatal day 4, P4), the surviving mutants showed a significant reduction in body weight and almost total lack of pigmentation (Fig. 1c). From P4 onwards, mutants additionally developed tremors, were largely unresponsive to stimuli, runted, whereas heterozygotes were still indiscernible from the wild-types. All mutants died within the first 10 days of life (Fig. 1d). Overall, these data suggest alterations of melanocytes and the peripheral nervous system (tremors) upon homozygous Adar1 deletion.
a Breeding strategy used to generate neural crest-specific Adar1 conditional knockout mice making use of the HtPA-Cre line. YFP allele is indicated in green; mutant genotype in blue. b Genotypes distribution and viability analysis at birth (postnatal day 0, P0). The number of animals of each genotype was compared to the expected ones to analyze viability. c Photos and body weight graphs of wild-types (WT, n = 11 at each stage), heterozygotes (Het, n = 3 at P0 and n = 4 at P4) and homozygous mutants (Mut, n = 7 at each stage) mice at P0 and P4. Note the significant body weight reduction at P4 and the total depigmentation of mutant mice. Asterisks represent p value: ***p < 0.001 determined using one-way ANOVA. d Survival rate (%) of wild-types (black), heterozygotes (gray) and mutants (blue) over time (days). Note that all mutants died within the first 10 days of life.
Absence of melanocytes in the skin of mutants after birth
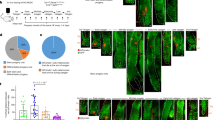
To determine the origin of the absence of pigmentation, we first performed a thorough histological analysis of the skin of wild-type, heterozygous (both referred as controls), and homozygous mutant mice, at P4. No specific alterations were evidenced, apart from the almost total absence of melanin from hair follicles of the mutants (Fig. 2a). RT-qPCR experiments on RNA extracted from dissected skin of controls and mutants revealed downregulation of the expression of melanoblasts markers (such as Sox10 and the melanocyte-specific isoform of Mitf) and melanocyte differentiation genes (Dct, Trpm1, and Tyr) in the mutants relative to controls (Fig. 2b). Use of the reporter R26R allele under the control of the Cre recombinase, to specifically mark NC cells and their derivatives on skin sections, additionally revealed a reduction of 96% in the percentage of hair follicles containing YFP-positive cells in mutants relative to controls (HtPA-Cre; Adar1fl/+) (Fig. 2c). In the very few mutant bulbs still containing YFP-positive cells, the number of these cells was also reduced although they produce melanin (Fig. 2d). Altogether, results suggest that lack of pigmentation upon Adar1 conditional deletion results from the lack of melanocytes, rather than from the absence of pigment within viable melanocytes.
a Paraffin skin sections from control and HtPA-Cre; Adar1fl/fl mutant mice stained with hematoxylin and eosin (H/E). Black arrowheads indicate presence of melanin in controls. Scale bar: 25 μm. b Relative expression level of melanocyte-expressed genes in skin of control (gray) and mutant (blue) mice (n = 5 or 8 controls and n = 4, 7 or 9 mutants depending on tested genes) analyzed by RT-qPCR. The relative abundance values of each amplicon were normalized to the internal control β-actin, and expression level of mutants and controls represented relative to controls. All data represent mean ± SEM, asterisks represent p value: ***p < 0.001 determined using t test. c Number (%) of follicles containing YFP-positive cells in the skin of control and mutant mice. Representative regions showing YFP (green) cells in follicles and quantification are presented in controls (gray) and mutants (blue). Statistical differences between the groups (n = 3 independent controls and mutants) were determined using t test (asterisks represent p value: ***p < 0.001). Scale bar: 50 μm. d Morphology and functionality of remaining YFP-positive cells (melanocytes) in follicles of control and mutant mice. Melanin synthesis (black) is shown on white light images. Bottom panels represent higher magnifications of upper panels (Scale bars: 50 and 15 μm, respectively). In (c) and (d) counterstaining with DAPI (blue) is shown to identify structures. Careful analysis of the few remaining YFP-positive cells in mutants skin showed them to be enlarged and able to produce melanin, suggesting that when still present, mutant melanocytes can retain at least part of their functionality. For (b) and (c), source data are provided as a source data file.
Based on these observations, we reanalyzed the Adar1 mutant pups at birth. Although not detectable by gross examination, RT-qPCR experiments (Fig. 3a, first graph) showed a drastic reduction of melanocyte-expressed genes in mutants relative to controls, thus suggesting that the defect is already present.
a–d Analysis of the expression of melanocytes differentiation genes (Sox10, Dct, Tyr) and Interferon-stimulated genes (ISG signature composed of Cxcl10, Isg15, Ifit1, Ifit2, Rsad2, Mx1) in the skin of controls (gray) and HtPA-Cre; Adar1fl/fl mutants (blue) at a P0, b, c E18.5 (note that we found mutant embryos of the two categories within the same litter) and d E16.5, by RT-qPCR. All data represent mean ± SEM. Statistical differences between the groups (for a n = 4 controls and n = 3–6 mutants depending on tested genes; for b n = 5 controls and n = 4 or 5 mutants depending on tested genes; for c n = 4 or 5 controls and n = 4–8 mutants depending on tested genes; for d n = 5 or 8 controls and n = 5 or 8 mutants depending on tested genes) were determined using t test (asterisks represent p values: *p < 0.05, **p < 0.01, ***p < 0.001). e YFP (green) and activated-caspase 3 (red) immunostaining on skin sections of control and HtPA-Cre; Adar1fl/fl mutant mice at E18.5 (white arrowheads indicate apoptotic cells) and quantification of caspase-3-positive cells among YFP cells (%). Data represent mean ± SEM, and statistical differences between the groups (n = 3 controls, gray and n = 6 mutants, blue) was determined using t test (asterisks represent p value: ***p < 0.001). Counterstaining with DAPI (blue) is shown to identify structures. Scale bar: 50 μm. For all panels, source data are provided as a source data file.
We next decided to breed the Adar1 model with mice expressing Cre recombinase driven by the Wnt1-promoter (triggering ablation in NC from E8.533). In this model, all the homozygous Adar1 mutant animals died within their first day of life for unknown reason. Nevertheless, RT-qPCR experiments and use of the YFP tracer showed alterations identical to the ones described above (Supplementary Fig. 1). Our data therefore collectively demonstrate that, irrespective of the Cre driver used, NC-specific deletion of Adar1 leads to a global lack of pigmentation, resulting from a drastic reduction in the number of melanocytes.
NC Adar1 deletion alters survival of melanocytes from E18.5
To determine the timing and cellular origin of defects observed, we performed similar RT-qPCR experiments at two embryonic stages (E16.5 and 18.5) and at birth to compare the expression of three melanocytic markers (Sox10, Dct, Tyr) (Fig. 3a–d; first column), and six ISGs (Cxcl10, Isg15, Ifit1, Ifit2, Rsad2, and Mx1, Fig. 3a–d second column), a signature recently shown to be increased upon Adar1 deletion in other tissues11. As previously mentioned, all the mutants showed marked downregulation of the expression of the three melanocytic genes tested at birth. This was concomitant to an elevated ISG signature relative to that of control littermates (Fig. 3a). At E18.5, half of the mutants showed downregulation of Dct and Tyr and strong upregulation of the ISG signature relative to controls (Fig. 3b). The others showed normal expression of Sox10, Dct, and Tyr, and a slight increase of ISGs (Fig. 3c). In contrast, there was no difference between the mutants and controls at E16.5 (Fig. 3d), a stage at which melanoblasts normally enter the developing hair follicle and start to differentiate22. Overall, these results suggest that the migration and differentiation of melanoblasts is not affected by Adar1 deletion up to E16.5. The disappearance of melanocyte-expressed genes occurs at approximately E18.5 and is preceded by upregulation of the ISG signature, leading us to analyze melanocytes survival at this embryonic stage.
Immunohistochemistry experiments using GFP and activated-caspase 3 antibodies revealed a high number of apoptotic YFP-positive cells in the epidermis and, to a lesser extent, in the hair follicles in the skin of mutants compared to controls at E18.5 (stainings and quantification, Fig. 3e), suggesting that the global depigmentation observed is due to the rapid loss of melanocytes by cell death. Of note, the number of CD45-positive cells was similar in the skin of mutants and controls at E18.5 and P4, suggesting that activation of ISGs and apoptosis occurs without immune cell recruitment (Supplementary Fig. 2).
NC Adar1 deletion alters SCs differentiation from birth
Based on the unsteady gate and tremors observed in the mutant animals, we speculated that Adar1 deletion may lead to the alteration of SCs development and/or myelin formation24. The sciatic nerves of mutants and controls were therefore analyzed, first at P4, by optical and electron microscopy (Fig. 4a–c). At the ultrastructural level, the control SCs had established a 1:1 relationship with axons and begun wrapping a myelin membrane along the axons, whereas the mutant ones failed to initiate myelination (Fig. 4b). Indeed, the axons of mutants were surrounded by cytoplasmic extensions of the SCs, showing no radial sorting alterations; but the SCs stalled at this promyelinating stage (no mesaxons, Fig. 4c). Quantifications confirmed a drastic decrease in the number of myelinated axon profiles per nerve section in mutants compared to controls, while the number of SCs and the axon diameters were similar (Fig. 4d). Both controls and mutants had Remak bundles, suggesting unaffected nonmyelinating SCs. Use of CD45 and (F4/80+CD11b+CD68) antibodies confirmed no, or rather limited, immune cells recruitment (Supplementary Fig. 3). Overall, these data suggest that Adar1 mutant SCs stall at the promyelinating stage, without evidence of cell degeneration.
a Toluidine blue-staining and b, c electron micrographs of transverse sections of sciatic nerves from control and HtPA-Cre; Adar1fl/fl mutant mice, illustrating the almost total absence of myelin in the mutants compared to controls. In mutants, the axons were surrounded by cytoplasmic extensions of the SCs, but the latter stalled at this promyelinating stage (b and insets shown in c). Note that a minority of mutant SCs myelinate axons properly (see b) along with absence of sign of axonal degeneration or B- or T-cell or macrophage invasion in the mutants compared to controls. Scale bars: 10 µm (a), 2 µm (b), 500 nm (c). d Quantifications performed from TEM pictures to count myelinated axons number per nerve, the SCs number per square millimeter and the distribution of axon diameter in controls (gray) and HtPA-Cre; Adar1fl/fl mutants (blue). Data of the three graphs represent mean ± SEM of n = 3 mice/group (Statistical differences between the groups were determined using t test (asterisks represent p value: ***p < 0.001, or Kolmogorov–Smirnov test, p values indicated on the graph). e DAPI (blue)/YFP(green), DAPI (blue)/MBP(green) or DAPI (blue)/Activated-Caspase 3 (red) immunostaining performed on sections of sciatic nerves from control and HtPA-Cre; Adar1fl/fl mutant mice along with the quantification of % of caspase-3-positive cells among YFP-positive cells. Data represent mean ± SEM. Statistical differences between the groups (n = 5 independent sections of four controls and n = 8 independent sections of five mutants) was determined using t test (asterisks represent p value: ***p < 0.001). Scale bar: 50 μm. f Relative expression of various SCs markers in sciatic nerve from control (gray) and HtPA-Cre; Adar1fl/fl mutant (blue) mice analyzed by RT-qPCR. All data represent mean ± SEM (n = 5 or 6 controls and n = 3 mutants depending on tested genes). Statistical differences between the groups were determined using t test (asterisks represent p value: ***p < 0.001). For (d–f), source data are provided as a Source data file.
These observations were consistent with immunohistochemistry experiments (Fig. 4e), which showed an apparently normal number of YFP-positive cells, an almost complete absence of MBP staining, but very few activated-Caspase3-positive cells in the sciatic nerves of mutants relative to controls (although statistically significant, percentage of apoptotic cells did not exceed 8% in controls and mutants, Fig. 4e). RT-qPCR experiments, performed on RNA extracted from isolated sciatic nerves, confirmed the reduced expression of several late SCs differentiation markers, such as Dusp15, Pmp22, Mpz, and Mbp in all mutants relative to controls. In contrast, the expression of earlier markers (Sox10 and Egr2) was not altered, confirming that the early development of these cells is not markedly altered by Adar1 deletion (Fig. 4f).
We next performed similar analyses on older mice (up to P10, latest stage available for Adar1 mutants) to determine whether Adar1 deficiency leads to a delay in or blockage of SCs differentiation. Semioptical and electron microscopy showed very little myelin in the mutants at P8 and P10 relative to controls (Supplementary Fig. 4a, b). Quantification again confirmed the drastic decrease in the number of myelinated axon profiles per nerve section, but no change in axon diameter at both stages (Supplementary Fig. 4c), suggesting a blockade of differentiation rather than a simple delay.
In parallel, we determined the onset of defects. Analysis of the expression of three SCs markers (Pmp22, Mpz and Mbp) and the ISGs signature revealed significantly greater ISGs expression in all mutants than controls from E18.5 (Supplementary Fig. 5b). This was followed by downregulation of the expression of the SCs markers from birth (Supplementary Fig. 5a), suggesting that SCs differentiation is blocked around birth and preceded by the upregulation of ISGs signature.
As with pigmentation, we confirmed these results using the Wnt1-Cre-mediated conditional deletion mouse model. At birth, electron microscopy showed a few myelinating SCs in controls but none in the mutants (Supplementary Fig. 6a). RT-qPCR experiments confirmed lower expression of Dusp15, Pmp22, Mpz, and Mbp in the mutants, but similar expression of Sox10 and Egr2 (Supplementary Fig. 6b). The early development of SCs is thus not altered by Adar1 deletion in either mouse line, but SCs appears to stall at or near promyelinating stage.
NC Adar1 deletion does not affect all NC derivatives equally
We determined if other NC derivatives were affected by Adar1 deletion. Analysis of three of these: enteric nervous system, heart structures derived from NC cells and craniofacial derivatives was performed. Macroscopic, histopathological analyses and Alcian blue followed by Alizarin red coloration revealed no obvious alterations of these three NC derivatives in mutants compared to controls at P4 with the techniques used (Supplementary Fig. 7a–c). However, 30% of the mutants (4 out of 13) present with cleft palate, suggesting some craniofacial alterations might be an incompletely penetrant feature.
RT-qPCR experiments on each tissue revealed increased ISGs expression in mutants relative to controls, but no alteration of NC expressed genes tested at P4 (Supplementary Fig. 7a–c). ISGs expression was however not increased in the liver of mutants (Supplementary Fig. 8), suggesting that this response is not systemic. Although we cannot exclude functional or cell-type-specific alterations not tested here, our results suggest Adar1 deletion does not affect all NC derivatives equally.
RNA-seq analysis in sciatic nerves of controls and mutants
To further define origin of Adar1 mutant SCs arrest and assess global changes in gene regulation, we performed transcriptomic analysis (RNA-seq) of sciatic nerves isolated from three control and three HtPA-Cre; Adar1fl/fl;R26R mutant animals at P4. The steady-state level of 3009 mRNAs differed by at least twofold between the two genotypes (Fig. 5a). We confirmed the results for a subset of these genes by RT-qPCR (Fig. 5b). The full list and the top 50-up and 50-downregulated genes are shown in Supplementary data 1 and Fig. 5c. Gene Ontology (GO) term and pathway analyses confirmed the profound enrichment of functions/pathways activated upon inflammation or after viral infection in the mutants versus controls (Fig. 5d, e). Screening of the 3009 differentially expressed RNAs against the interferome database34 showed that 2253 (75%) are known to be regulated by IFN type 1 or 2 (Fig. 5f). Aside from activation of the innate immune response, the GO terms analysis also showed activation of other biological functions in mutants relative to controls, including growth-factor activity and positive regulation of the epithelial to mesenchymal transition (EMT) (Fig. 5d). The latter being critical for NC development, we looked at expression of key EMT-related genes in our dataset. A decrease in Cdh1 expression (E-cadherin), and upregulation of the master regulators Twist1 and 2, Snail2, Lef1, and Met were observed (Supplementary data 1). However, the expression of well-known mesenchymal markers Cdh2 (N-cadherin) and Vimentin were not altered, suggesting that only part of the molecular machinery associated to EMT is activated upon Adar1 deletion.
a Volcano plot of differentially expressed genes in sciatic nerves of HtPA-Cre; Adar1fl/fl mutants compared to controls (upregulated in mutants: red; downregulated in mutants: green; unchanged in black). Results were considered statistically significant for p values ≤ 0.05 and fold-changes ≥ 2.0 (n = 3 for each condition) using Deseq2 R package for differential analyses. In all, 1664 were upregulated in the mutant versus control animals and 1345 were downregulated, suggesting ADAR1 is involved in mechanism governing gene repression and activation. b Validation of a subset of differentially regulated genes on RNA extracted from isolated sciatic nerves from controls and mutants by RT-qPCR. Relative expression levels in controls (gray) and mutants (blue) are presented as mean ± SEM. Statistical differences between the groups (n = 4 controls and n = 4 mutants) were determined using t test (asterisks represent p values: *p < 0.05, **p < 0.01, ***p < 0.001). Source data are provided as a source data file. c Heatmap representation of RNA-seq analysis depicting the top 50-up and 50-downregulated genes in mutants relative to controls. Colors reflect the z-score. d Representation of enriched functions by GO term and enriched signaling pathways (using GO analysis and Ingenuity). e Functions/signaling pathways enriched in mutants versus controls are indicated in red, those enriched in controls versus mutants are indicated in light green. f The interferome database V2.01I [http://interferome.its.monash.edu.au/interferome/] was used to determine the number of interferon regulated genes (IRGs) specific to different types of Interferon IFN (I, II and III) within the 3009 differentially regulated genes upon Adar1 deletion. Overlap of 75% (2253/3009) was evidenced. Fisher’s exact test confirmed enrichment (p < 2.2E-16). g Heatmap representation of differentially expressed transcriptional regulators (found in association with GO terms: DNA-templated transcription, positive or negative regulation of transcription from RNA polymerase II promoter, positive or negative regulation of transcription, DNA-templated transcription factor activity, sequence-specific DNA binding, and positive regulation of gene expression). Out of the 281 factors identified, 69 were deregulated more than fivefold in mutants relative to controls. Name of each gene is indicated on the left, along with its belonging to the Interferome dataset. Colors reflect the z-score.
Consistent with myelination alterations, genes encoding myelin proteins were among the top 50-downregulated genes in the Adar1 mutants relative to controls (Fig. 5c and Supplementary data 1). Additionally, cholesterol biosynthesis and metabolism (one of the main components of myelin sheaths) and lipid metabolism were among the processes significantly enriched in controls relative to the mutants (Fig. 5d, e).
Based on the alterations of SCs observed (block at promyelinating stage), we next analyzed the differential expression of key genes (transcription factors in particular) known to control this specific step of development26,30. Among these, expression of Sox10, Egr2, Egr3, yy1, Sox2, Jun, Id2, Pax3, and Nfkb1 was not significantly altered in the Adar1 mutants relative to controls. Of note, the expression of Egr1, which is normally downregulated at the promyelinating stage35, was significantly higher in the Adar1 mutants (12.5-fold increase, Supplementary data 1). In contrast, the expression of Pou3f1 and Pou3f2, known to be transiently expressed in SCs26,36 and peaking at the promyelinating stage, were both lower (ratios of 0.23 and 0.44, respectively). The deregulation of the expression of these three candidate transcription factors may be a direct consequence of, contribute to, or be responsible for the observed promyelination blockade of differentiation.
To determine whether additional factors involved in transcriptional regulation are also deregulated, we examined the relative expression of other genes associated with transcriptional regulation (see Methods for the GO term used). Among these, 69 were deregulated more than fivefold in mutants relative to controls. Most of them were upregulated in the mutants (64/69) and 87% were found within the interferome database (Fig. 5g). Among them, we again noticed the presence of Egr1. There was also increased expression of Hes1, Myc, Tfap2a, and Tfap2b, previously shown to be negative regulators of myelination or transcription factors known for their function in early NC development26,37. In addition to Hes1, the Notch effector Hey2 was also upregulated in mutants (4.7-fold-change, Supplementary Data 1), arguing for persistently activated inhibitory Notch signaling in Adar1 mutants relative to controls. Overall, data suggest that SCs developmental arrest in Adar1 mutants could be due to the persistent expression of limited number of these regulators of myelination.
Injury-associated genes activation in Adar1 mutants
Intriguingly, the top 50 upregulated genes included several known to be activated upon nerve injury25,38. In particular, Shh and Gdnf, two markers that are not expressed in SCs in normal mature or developing nerves, but known to be activated SCs during repair, were markedly upregulated in the mutant mice (Fig. 5c). We therefore evaluated the extent of the overlap between the two conditions by comparing our RNA-seq data to several lists of mRNAs previously shown to be deregulated 1, 5, or 7 days after nerve injury39,40. Strikingly, 34% of the mRNAs deregulated upon Adar1 deletion are also deregulated upon nerve injury, including decreased expression of myelin genes and those for cholesterol biosynthesis, increased expression of a subset of early SC markers, activation of genes involved in the innate immune response, and increased expression of specific repair markers, such as Shh, Gdnf, Artemin, Fgf5, Wif1, and Lgals325,41,42 (Fig. 6a and Supplementary data 2 for the 1028 overlapping mRNAs). The differential expression of some of these was further validated by RT-qPCR (Fig. 6b). Because the morphological transition from differentiated/mature SCs into a repair SCs was also shown to be associated with an EMT signature39, we compared the datasets. Of the 111 EMT-associated genes found to be differentially expressed between uncut and cut nerves39, 36% were also deregulated upon Adar1 deletion (Supplementary Table 1 and Fig. 6c). Overall, analysis of the RNA-seq dataset suggests that activation of a program resembling response to nerve injury occurs upon conditional NC deletion of Adar1.
a Venn diagram showing significant overlap between RNA-seq data generated in this study (orange) and differentially expressed genes upon peripheral nerve injury (blue)39,40. The number of genes in each category, the % of overlap and representative genes are indicated. Fisher’s exact test confirmed enrichment (p < 2.2E-16). Note that 1028 out of 3009 genes we found deregulated are overlapping with published datasets. b Validation of a subset of differentially expressed genes by RT-qPCR. All data represent mean ± SEM. Statistical differences between the groups (n = 4–6 controls and n = 4–8 mutants depending on tested genes) were determined using t test (asterisks represent p values: *p < 0.05, **p < 0.01, ***p < 0.001). Source data are provided as a source data file. c Of the 111 EMT-associated genes found to be differentially expressed between uncut and cut nerves39, 34% were also deregulated upon Adar1 deletion. Top 10 up-(pink) and down-(green) regulated genes found in common, (p-adj < 0.05) are indicated. Note that “EMT-associated genes” are more generally associated to migration and cell adhesion function.
A-to-I RNAs targets and Mda5 shutdown rescue experiments
We next wished to directly determine the contribution of A-to-I editing. Making use of the pipeline described in Methods, we identified 60 A-to-I (G) editing sites specifically altered in mutants within or near 57 different genes in our RNA-seq dataset (Supplementary Data 3). Only six of these sites were previously reported in the RADAR or DARNED databases, suggesting that most of the identified sites may be regulated in a tissue-specific manner. RNA editing sites led to amino-acid change, intronic, splice acceptor site or 3′UTR variants with similar frequency. Only seven of the 57 targets showed deregulation of over twofold. All seven targets were in the interferome database, while only three were previously associated to repair: Atp8b1, Gusb, and Slc7a11. We also examined the known function of each of the 57 associated genes. Some (Ufc1 and Ctf3c3) are involved in disorders with CNS myelin alterations (OMIM 618076). NC deletion of Ilk leads to alteration of SCs myelination43. Nevertheless, expression and splicing of these genes were similar in the mutants and the controls upon bioinformatics analysis. Slc7a11 is the only gene found to be deregulated and with a probable association with myelin production.
Irrespective of the identity of the targeted RNA, it seemed likely that alterations of A-to-I mismatches could trigger chronic activation of ISGs after their recognition by MDA5 as in other tissues10. We therefore tested whether concomitant deletion of Ifih1 (encoding Mda5) could rescue the SCs defects observed. In primary cultures of mixed neurons and SCs under enhanced myelinating conditions44, we first observed that MBP-positive segments (mark of myelination) were absent from the neurons (labeled by TUJ1) of mutants while present in controls (Fig. 7a). As in vivo, these defects were accompanied by upregulation of the expression of the two tested ISGs relative to controls (Cxcl10 and Isg15, Fig. 7b). Strikingly, we observed MBP-positive myelin segments in Adar1 mutants transduced with Sh-Ifih1 (>60 segments per cultures), but none in those transduced with the Sh-control (Fig. 7a). This rescue was concomitant with a return of the two ISGs expression to basal levels (Fig. 7b). Altogether, our results show that Adar1 safeguards SCs from unwanted MDA5-mediated ISGs activation, a process that leads to the alteration of myelin formation if deregulated.
a TUJ1 (green)/MBP (red) immunostaining performed on primary cultures of mixed neurons and SCs from dorsal root ganglia (DRG) of control and HtPA-Cre; Adar1fl/fl mutant embryos at E13.5. Cultures were untransduced (no infection, Ctl) or transduced with lentivirus containing Sh-control (Sh-ctl) or Sh-Ifih1 just before myelination induction. Note the absence of MBP in Adar1 mutants and the restored phenotype upon Sh-Ifih1 transduction. Counterstaining with DAPI (blue) is shown to identify cell density. Scale bar: 12 μm. b Relative mRNA expression of Ifih1 (Mda5, to show Ifih1-shRNAs efficacy validation), and two ISGs (Cxcl10 and Isg15) analyzed by RT-qPCR on RNAs extracted from untransduced (−) and transduced primary cultures of controls (gray) and mutants (blue). All data represent mean ± SEM. Statistical differences between the groups (n = 4 controls and n = 4 mutants) were determined using t test (asterisks represent p values: *p < 0.05, **p < 0.01, ***p < 0.001). c Pigmented melanocytes (melanin-producing cells) observed after culture of NC cells issued from trunk region of neural tube of control and Adar1 mutant embryos at E9.5. No discernable differences could be observed between controls and mutants during the first 7 days of culture, but note the reduction of melanin-producing cells in Adar1 mutants (n = 7) compared to controls (n = 6) after 21 days of culture. Three days after neural tube explant deposition on fibronectin, cultures were untransduced (Ctl) or transduced with lentivirus containing Sh-Ifih1. Note the restored phenotype of mutants upon Sh-Ifih1 transduction (n = 5). Sh-Ifih1 transduction had no discernable effects on control cultures (n = 3). Scale bar: 100 μm. Source data are provided as a source data file.
On the same line, we tested whether concomitant deletion of Ifih1 could rescue the melanocytes defects observed. Trunk neural tube explants cultures from E9.5 embryos performed under conditions favoring melanocytes proliferation and differentiation revealed a drastic reduction in the number of melanin-producing melanocytes in mutants compared to controls (Fig. 7c, ctl line, mean ± SEM of 167 ± 42 and 13 ± 12 melanocytes counted in six controls and seven mutants respectively,***p < 0.001, t test). As for SCs, Ifih1-ShRNAs infection increased the number of melanin-producing cells in Adar1 mutants (49 ± 12, **p < 0.01; Fig. 7c), suggesting that concomitant deletion of Mda5 at least partially rescue pigmentation defects. In light of these results, the phenotype of Adar1;Mavs11 and Adar1;Ifih1 double mutants (ref. 45 and D. Stetson, personal communication) was re-interpreted and revealed no pigmentation anomalies in either models, suggesting that pigmentation defects we observed in vitro and in vivo result from a mechanism that is MDA5/MAVS dependent.
Discussion
We used a conditional deletion strategy in mice to study the role of Adar1 and A-to-I RNA editing during NC development. The first alteration we observed in mutants was global depigmentation. This was true irrespective of the NC-specific Cre driver line used (HtPA-Cre or Wnt1-Cre). RT-qPCR experiments using various melanocytic markers and use of the R26R allele under the control of the NC-specific Cre recombinase to trace YFP-positive melanocytes revealed that the early steps of melanoblast development are unaffected. However, there is a severe reduction in the number of melanocytes within the hair follicles of mutants from E18.5, resulting from the rapid death of these cells, and following upregulation of an ISGs signature recently shown to be activated upon Adar1 deletion/RNA editing activity alteration in other tissues. As when Adar1 is deleted in hematopoietic and neuronal progenitors, massive cell death of melanocytes therefore explains the depigmentation observed1,5,7.
NC Adar1 deletion also severely alters SCs differentiation, i.e. no myelin is formed. Again, this observation was made using both NC-specific Cre drivers. Electron microscopy, RT-qPCR, and RNA-seq analyses showed that the mutant SCs are blocked at the promyelinating stage. Limited impact on SCs number and no difference in axonal diameter were observed, suggesting no evident alterations of neurons. This phenotype recalls that of Egr2, Dicer, and Dgcr8 mutants42,46,47,48, in which most of the SCs stall at the promyelinating stage, are unable to myelinate, and some radial sorting defects are observed. In Dicer and Dgcr8 mutants, artificially sustained expression of genes characteristic of immature/promyelinating cells is proposed to explain the defects. Both mutants indeed display decreased Egr2 expression and increased expression of Sox2, Jun and Notch, Hes1, Egr1, Egr3,Ngfr, Ncam1, and Ccnd142,46,47,48. In our case, the expression of these genes was not significantly altered, except for Egr1 and Hes1, which were upregulated in the Adar1 mutants relative to controls. Although the phenotypes are very similar, these observations suggest that the underlying molecular mechanism is different. In the mutants presented here, additional deregulated expression of Pou3f1, Pou3f2, Tfap2a, Tfap2b, Myc, and Cebps transcription regulators known to play key roles in SCs or early NC development was noticed. Whether their deregulation is a cause or a consequence of the observed alterations is under investigation.
Interestingly, 34% of the RNA transcripts deregulated upon Adar1 deletion are also deregulated upon nerve injury39,40. Injury-induced SCs reprogramming involves the downregulation of myelin genes and upregulation of immature SCs markers. In our case, however, some (but not all) early markers were more likely maintained and the myelin genes not induced. An additional specific molecular signature appears during injury, including (1) upregulation of neurotrophic factors promoting survival of the injured neurons; (2) upregulation of the expression of cytokines as part of the innate immune response; and (3) activation of a cell-intrinsic myelin breakdown process (autophagy)25. We also observed increased expression of Shh, Gdnf, Nrg1 Artemin, Fgf5, Vgf, Btc,Tnf, and Edn1 in Adar1 mutants. Because some of these (e.g. Nrg1 and Edn1) are known to support SCs survival24,49,50, this upregulation may explain why Adar1 mutant SCs escape the massive cell death encountered in melanocytes. In our case, the inflammatory response was triggered by A-to-I RNA editing alterations (see below), but no or limited recruitment of immune cells was observed. Of note, Dgcr8 mutants, but not Egr2 mutants, also show upregulation of an injury-related gene expression program42. In addition to epigenetic mechanisms28,38,39, RNA editing could therefore be involved in the control of injury-related gene expression programs. As for the Dicer and Dgcr8 mutants, it would be of specific interest to test the function of ADAR1 in myelin maintenance under standard conditions or stress paradigms.
As in other tissues, analysis of the differentially expressed mRNAs using the interferome database additionally showed 75% of them to be within or associated with IFN type 1 or 2 signaling pathways34. However, analysis of liver from mutant mice revealed ISGs signature is not systemic, suggesting that activation of at least part of these ISGs is intrinsic to/specific for NC derivatives. SCs and melanocytes, which are typically nonimmune cells, indeed possess the capacity to elicit innate immune response by expressing a large number of innate immune genes in response to infections51,52. It is however difficult to ascertain the causality of such a large number of genes and identify primary versus secondary events in the phenotype genesis. Nevertheless, some of these ISGs are early NC expressed genes or negative regulators of myelination, and/or are part of the injured response described above, suggesting that chronic activation of some of them may dysregulate the balance between positive and negative regulators of myelination, explaining the observed blockade of myelination. Of interest, 61 genes known to be transiently expressed after injury are abnormally maintained in mutants with a fold-change > 5, including Soat2, Vgf, Egr1, Ch25h, which have known functions in myelination/demyelination processes (Supplementary Data 2, genes in red). Their deregulation could therefore participate to the phenotype genesis.
Sixty A-to-I (G) editing sites were also specifically altered in mutants within or near 57 different genes. Whereas genome-wide analysis of the in vivo substrates of ADAR1 previously led to the identification of clustered editing within long dsRNA stem loops within the 3′ untranslated regions of endogenous transcripts9,10, no such enrichment was observed here. Association between each of the concerned genes and SCs alterations seems also very limited. Although the identification of unedited RNA at the origin of any of the observed phenotypes will require further validation, we propose that their recognition by MDA5 is (as in other tissues) part of the pathogenic pathway10. Indeed, (i) extinction of Ifih1 partially rescued the differentiation of Adar1 mutant SCs in vitro, (ii) Sh-Ifih1 at least partially rescued melanocytes defects of Adar1 mutants in vitro, (iii) reinterpretation of the phenotype of Adar1;Mavs and Adar1;Ifih1 double mutants in light of our results revealed no pigmentation anomalies (refs. 11,12,45 and D. Stetson, personal communication).
Although both Cre lines used allow deletion of Adar1 in a large number of NC derivatives, it is striking that the alterations mainly affect melanocytes and SCs (no major or incomplete penetrant alterations of other NC structures identified to date) and could be observed only from E18.5 onwards. The trans-regulators that trigger the deregulation of RNA editing or the activation of cell-specific RNA species that initiate MDA5-mediated IFN production from E18.5 remain to be identified.
Beyond the description of the mouse phenotype, our results could help further extend the clinical spectrum associated to ADAR1 mutations in human. Only one patient with AGS due to ADAR1 mutation has been reported to present with associated peripheral neuropathy13. Data presented here suggest that neuropathy could have been overlooked considering the extreme disability of most patients. An oriented neurological evaluation of patients with AGS syndrome and ADAR1 mutations is therefore needed to answer this question.
Our study also further expands our knowledge of the processes that drive DSH in humans16,19,53,54. Although the number of DSH patients with ADAR1 mutations is increasing, very few studies have focused on the origin of the observed alterations19,53,54. Two reports showed a decreased number of melanocytes in hypo-pigmented regions19,55 along with immature melanosomes, numerous vacuoles and signs of degenerative mitochondria53. We show here that the depigmentation identified upon Adar1 deletion is due to a massive reduction in the number of melanocytes through apoptosis. Our results also recall observations made in the hyper-pigmented regions of DSH patients. In these regions, the melanocytes were reported to be enlarged, containing numerous and elongated dendrites, said to increase their functionality. In the present mouse model, the very few surviving Adar1 mutant melanocytes were also enlarged relative to controls and produce melanin (legend of Fig. 2d). Whether their functionality is modified is yet to be determined. Despite these overall similarities, timing of the pigmentation defects seems to differ between species (depigmentation during infancy in humans versus loss of melanocytes before birth in mice). However, recent publications suggest that ADAR1 mutations can also be associated with DSH from birth56. Aside DSH, similar findings are frequently observed in other inflammation-associated pigmentary disorders, including vitiligo57, opening the possible involvement of ADAR1 and alterations of A-to-I editing in this disorder.
In conclusion, we highlight the function of ADAR1 in the development of melanocytes and SCs. Overall, our results suggest that this enzyme safeguards these NC-derived cells from unwanted MDA5-mediated IFN production and chronic ISG upregulation, with implications for the human diseases caused by ADAR1 mutations, NC development and NC-linked-disorders.
Methods
Mice, genotyping and global phenotype analysis
The mouse models used in this study were: (i) B6.129-Adartm1knk/Mmjax, a model purchased from the Jackson Laboratory (stock number 34619-JAX, here referred as Adar1fl/fl), (ii) Gt(ROSA)26Sortm1(EYFP)Cos32 (referred as R26R); (iii) Tg(PLAT-Cre)116Sdu31 (referred as HtPA-Cre), and (iv) Wnt1-Cre driver33.
Cre-floxed systems for gene deletion were used to get animals of interest. Breeding of Adar1fl/fl with R26R was first performed to obtain animals homozygous at both loci. Those were subsequently crossed with HtPA-Cre or Wnt1-Cre drivers to generate HtPA-Cre;Adar1fl/+;R26R or Wnt1-Cre;Adar1fl/+;R26R animals. Each of these were then crossed with Adar1fl/fl;R26R to generate progenies of different genotypes (HtPA-Cre; Adar1fl/fl;R26R or Wnt1-Cre; Adar1fl/fl;R26R) referred as mutants; wild-type (Adar1fl/fl;R26R or Adar1fl/+;R26R) and heterozygous for Adar1 (HtPA-Cre; Adar1fl/+;R26R or Wnt1-Cre;Adar1fl/+R26R). Tails biopsies were used to perform DNA extraction making use of the direct PCR lysis reagent for Tail (Viagen) and subsequent genotyping. All primers purchased from Eurogentec and PCR primers are reported in Supplementary Table 2. The study complies with all relevant ethical regulations for animal testing and research. Indeed, experiments were done in accordance with the Institutional Animal and Use Committee guidelines of the Institut National de la Santé et de la Recherche Médicale (INSERM). The study received ethical approval from the Comite d’Ethique pour l’experimentation Animale (C2EA-12-035 and 16-097) and deposited under APAFIS 9783 number 2017022215395397.
Histology and electronic microscopy
Tissues (sciatic nerves, skin, gut and heart) were dissected and fixed in 4% paraformaldehyde, paraffin-embedded cuts and stained with hematoxylin and eosin. Alternatively, fixed tissues were frozen in Optimal Cutting Temperature media (OCT, Sakura), cut into 8 μm sections and used for immunofluorescence experiments. Skeletal preparations were performed on P4 controls and mutant embryos by Alcian Blue and Alizarin red. Briefly, the mice at P4 were eviscerated, fixed overnight in ethanol and then macerated in acetone for 24 h. The fixed fetuses were stained with Alcian Blue 8GX (150 mg/l in 80% ethanol, 20% acetic acid) for 48 h at 37 °C, washed in 70% ethanol for 8 h and cleared in 1% KOH overnight. They were then incubated in Alizarin Red S (50 mg/l in 2% KOH) for 6–8 h, cleared in 1% KOH and stored in 50% glycerol before observation.
For electron microscopy, mutant and control sciatic nerve samples were collected and fixed at 4 °C in 2.5% glutaraldehyde in sodium cacodylate buffer, rinsed and post fixed at 4 °C in a 1% osmium tetroxide solution. Samples were then dehydrated in graded alcohol and acetone baths and embedded in epoxy resin. After polymerization (for 48 h at 60 °C), samples were removed. Semi-thin transverse sections (1.5 μm) were stained with toluidine blue and ultrathin sections were stained with uranyl acetate and lead citrate and observed through a JEOL electron microscope. The number of myelinated nerve fibers in each nerves was counted manually on semi-thin transverse sections. The SCs count was performed manually based on photographs of nerve biopsy (electron microscopy) of control and mutant mice at each stage of development and reported to the surface studied to determine a density of cells per mm2. On these same EM pictures, after having identified myelinated nerve fibers (and nerve fibers in the process of myelination), the size of their axons was measured for each of them using ImageJ freeware (National Institutes of Health, USA).
Immunofluorescence and immunohistochemistry
Frozen sections were blocked in PBS + 0.1% Triton X-100 (PBT) for 1 h. Primary antibodies were diluted in blocking solution (PBT + 1% BSA + 0.15% glycine) as follows: cleaved-Caspase-3 (casp3, D175, lot 43, rabbit, Cell Signaling Technologies, 1:200 dilution), Mbp (SMI 94R-0100 lot E10172EF, mouse, Covance, 1:1000 dilution), GFP (A11122, lot 1891900, rabbit, Invitrogen, 1:1000 dilution), CD45 (14-045-185 C30F11, Rat, Invitrogen, 1:100 dilution) and F4/80, CD11b, CD68 cocktail (MCA497, MCA74G, MCA1957, Rat, Bio-Rad, 1:200 dilution) and incubation performed overnight at 4 °C. After several washes, slides were incubated with secondary antibody (anti-rabbit alexaFluor555, anti-mouse alexaFluor488, anti-rat Cy3 or anti-rabbit alexaFluor488, Invitrogen, 1:500 dilution each) for 2 h before final washing. For dorsal root ganglia (DRGs) cultures, cells were fixed in 4% PFA for 10 min and immunostained using the same procedure with TuJ1 (MMS-435P, lot TU17044, mouse, Eurogentec, 1:1000 dilution) and Mbp (ab40390, Rabbit, Abcam, 1:200 dilution). Secondary antibodies used were as follows: anti-mouse alexaFluor 488 and anti-rabbit alexaFluor 555 (Invitrogen, 1:500 dilution each). Preparations were then mounted using Vectashield containing DAPI (Vector laboratories) and observed using Spinning Disk from Zeiss.
Quantitative real-time PCR
Skin, sciatic nerves, heart, gut, mandible and liver from P4 and newborn animals or E16.5 and E18.5 embryos of different genotypes were dissected and total RNA extracted using RNeasy Mini or Micro kit (Qiagen) according to the manufacturer’s instructions. After DNAse treatment, concentration and quality of RNA was determined using the NanoDrop (Thermofisher) and Xpose (Trinean) apparatus. cDNA was synthesized using the Maxima First Strand cDNA Synthesis. Quantitative real-time PCR (qPCR) was performed using the Maxima SYBER green/ROX qPCR Master Mix X2 (all from Thermofisher) and amplified using Mastercycler® RealPlex2 (Eppendorf). The relative abundance values of each amplification product were normalized to the internal control β-actin, and mRNA expression levels in mutants expressed relative to controls. PCR primers are reported in Supplementary Table 3.
RNA-seq library preparation and sequencing
Total RNAs of sciatic nerves were isolated using the RNeasy Kit (Qiagen) including a DNAse treatment step. RNA quality was assessed using RNA Screen Tape 6000 Pico LabChips with the Tape Station (Agilent Technologies) and the RNA concentration was measured by spectrophometry using Xpose. RNA-seq libraries were prepared starting from 60 ng of total RNA using the Universal Plus mRNA-Seq (Nugen) as recommended by the manufacturer. The oriented cDNA produced from the poly-A + fraction were PCR amplified (15 cycles). An equimolar pool of the final indexed RNA-Seq libraries was sequenced on an Illumina HiSeq2500 (paired-end reads 130 bases + 130 bases). A mean of 124 millions of passing filter paired-end reads was produced per library sample (with a minimum of 103 million and a maximum 158 millions).
RNA-seq alignment and bioinformatics data analysis
RNA-Seq data analyses were performed by GenoSplice [https://www.genosplice.com/]. Sequencing, data quality, reads repartition (e.g., for potential ribosomal contamination), and insert size estimation were performed using FastQC, Picard-Tools, Samtools and rseqc. Reads were mapped using STARv2.4.058 on the mm10 Mouse genome assembly. Gene expression regulation study was performed following classic scheme59,60,61. Briefly, for each gene present in the Mouse FAST DB v2018_1 annotations, reads aligning on constitutive regions (that were not prone to alternative splicing) were counted. Based on these read counts, normalization and differential gene expression were performed using DESeq262 on R (v.3.2.5). Only genes expressed in at least one of the compared experimental conditions were further analyzed. Genes were considered as expressed if their rpkm value was greater than 97.5% the background rpkm value based on intergenic regions. Results were considered statistically significant for p values ≤ 0.05 and fold-changes ≥ 2.0 in mutants compared to controls. 20.1% of the expressed genes (3009/14402) were thus found up- or downregulated in mutants versus controls. GO term and pathway analysis were subsequently performed on this list, making use of KEGG, REACTOME, and Ingenuity pathway analysis. We sought A-to-I (G) RNA variants using union of results from GATK HaplotypeCaller and RED-ML63 on minimal total coverage in all samples ≥ 10 reads. A to G RNA variants were annotated using VEP and filtered if they correspond to already known genomic variant from dbSNP142, and retained if % editing level was significantly reduced in mutants compared to control samples (p ≤ 0.1). To check if editing sites were previously reported, we used the RADAR or DARNED databases [https://rnaedit.com] and [https://darned.ucc.ie]. Several of these were found similarly altered in independent set of four control and mutant samples.
Mixed SCs and NC cultures and rescue experiments
DRGs cultures were prepared from isolated DRGs of E13.5 embryos. Briefly, dissociated cells were plated at a density of 4 × 104 cells per 13 mm coverslip coated with Matrigel (BD Biosciences) and poly-d-Lysine (Sigma). Cells were kept in neurobasal medium supplemented with B-27, penicillin/streptomycin, glutamax (from Gibco life Technologies) and 50 ng/ml NGF (PreproTech) for 2 days, and then in basal medium supplemented with glutamax, d-glucose, BSA, P/S, ITS and NGF for seven additional days as in ref. 44. Cells were then maintained for 10–13 additional days in medium supplemented with 50 µg/ml L-ascorbic acid (Sigma), 15% heat inactivated calf serum and 5 µg/ml heparin (Sigma) to induce myelination, before fixation.
Neural tube explants were obtained from E9.5 embryos and plated on six-well plates coated with 20 µg/ml of human plasma fibronectin (GIBCO BRL) diluted in PBS. The basal culture medium consisted of F12 medium supplemented with 10% fetal calf serum, 16 nM TPA (Sigma), 2 μg/ml [Nle4, D-Phe7]-a-MSH (Sigma) and Edn3 at a concentration of 3 nM64. Medium was changed every second to third day for a period of 21 days to allow melanocytes differentiation and melanin production before fixation.
Three days after neural tube explants seeding or before myelination induction, cells were infected with pLKO ShRNA targeting Ifih1 (TRCN0000103646: 5′-CCACAGAATCAGACACAAGTT-3′) or control ShRNA (SHC002V) (Sigma) with a multiplicity of infection (MOI) of 2. Infection was performed using standard procedure. We chose this strategy because lentiviral ShRNA-mediated KO is efficient in SCs and this specific Sh-Ifih1 was previously used with success in Adar1 mutant hematopoietic stem and progenitor cells10.
Statistical analysis
All analyses were done using Graphpad Prism 6.0. Data are presented as mean ± SEM in the graphs and were analyzed using unpaired two-tailed Student’s t test for simple comparison or one-way ANOVA for multiple comparisons. p < 0.05 was considered statistically significant. p values were as follows: not significant (ns) >0.05, *<0.05, **<0.01, ***<0.001. Fisher Exact test was used to delineate data overlap presented in Venn diagrams of Figs. 5 and 6. For distribution plots presented in Fig. 4 and Supplementary Fig. 4, the Kolmogorov–Smirnov test was used for statistical analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The data that support the findings of this study are all available within the text, supplementary, or source data files. The source data underlying Figs. 2b, c, 3a–e, 4d–f, 5b, 6b, 7b, c, and Supplementary Figs. 1a, 2, 3, 4c, 5a–c, 6b, 7a–c, 8 are provided as a Source Data file. The RNA-seq datasets generated during the study have been submitted to the [GEO] repository and accepted under [GSE127795]. The datasets reanalyzed for comparison purpose are [E-MTAB-5633] in Array Express [https://www.ebi.ac.uk/gxa/experiments/EMTAB-5633?ref=aebrowse] and [GEO] database [accession #GSE33454] provided in refs. 39,40.
References
Walkley, C. R. & Li, J. B. Rewriting the transcriptome: adenosine-to-inosine RNA editing by ADARs. Genome Biol. 18, 205 (2017).
Nishikura, K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 7, 919–931 (2006).
Tan, M. H. et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017).
Hogg, M., Paro, S., Keegan, L. P. & O’Connell, M. A. RNA editing by mammalian ADARs. Adv. Genet. 73, 87–120 (2011).
Walkley, C. R., Liddicoat, B. & Hartner, J. C. Role of ADARs in mouse development. Curr. Top. Microbiol. Immunol. 353, 197–220 (2012).
Higuchi, M. et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406, 78–81 (2000).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem 79, 321–349 (2010).
Hartner, J. C. et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 279, 4894–4902 (2004).
Liddicoat, B. J. et al. Adenosine-to-inosine RNA editing by ADAR1 is essential for normal murine erythropoiesis. Exp. Hematol. 44, 947–963 (2016).
Liddicoat, B. J. et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349, 1115–1120 (2015).
Pestal, K. et al. Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43, 933–944 (2015).
Mannion, N. M. et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 9, 1482–1494 (2014).
Crow, Y. J. et al. Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am. J. Med. Genet. A 167A, 296–312 (2015).
Rice, G. I. et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 44, 1243–1248 (2012).
Rice, G. I. et al. Genetic, phenotypic, and interferon biomarker status in ADAR1-related neurological disease. Neuropediatrics 48, 166–184 (2017).
Hayashi, M. & Suzuki, T. Dyschromatosis symmetrica hereditaria. J. Dermatol. 40, 336–343 (2013).
Miyamura, Y. et al. Mutations of the RNA-specific adenosine deaminase gene (DSRAD) are involved in dyschromatosis symmetrica hereditaria. Am. J. Hum. Genet. 73, 693–699 (2003).
Slotkin, W. & Nishikura, K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 5, 105 (2013).
Sheu, H. M. & Yu, H. S. Dyschromatosis symmetrica hereditaria—a histochemical and ultrastructural study. Taiwan Yi Xue Hui Za Zhi 84, 238–249 (1985).
Larue, L., de Vuyst, F. & Delmas, V. Modeling melanoblast development. Cell Mol. Life Sci. 70, 1067–1079 (2013).
Le Douarin N. M., & Kalcheim C. The Neural Crest (Cambridge University Press, 1999).
Mort, R. L., Jackson, I. J. & Patton, E. E. The melanocyte lineage in development and disease. Development 142, 1387 (2015).
Nave, K. A. & Werner, H. B. Myelination of the nervous system: mechanisms and functions. Annu. Rev. Cell Dev. Biol. 30, 503–533 (2014).
Salzer, J. L. Schwann cell myelination. Cold Spring Harb. Perspect. Biol. 7, a020529 (2015).
Jessen, K. R. & Mirsky, R. The repair Schwann cell and its function in regenerating nerves. J. Physiol. 594, 3521–3531 (2016).
Stolt, C. C. & Wegner, M. Schwann cells and their transcriptional network: Evolution of key regulators of peripheral myelination. Brain Res. 1641, 101–110 (2016).
Levy, C. et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell 141, 994–1005 (2010).
Ma, K. H. & Svaren, J. Epigenetic control of Schwann cells. Neuroscientist 24, 627–638 (2018).
Mirsky, R. et al. Regulation of genes involved in Schwann cell development and differentiation. Prog. Brain Res. 132, 3–11 (2001).
Salzer, J. L. Switching myelination on and off. J. Cell Biol. 181, 575–577 (2008).
Pietri, T., Eder, O., Blanche, M., Thiery, J. P. & Dufour, S. The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev. Biol. 259, 176–187 (2003).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Danielian, P. S., Muccino, D., Rowitch, D. H., Michael, S. K. & McMahon, A. P. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 8, 1323–1326 (1998).
Rusinova, I. et al. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41, D1040–1046 (2013).
Topilko, P. et al. Differential regulation of the zinc finger genes Krox-20 and Krox-24 (Egr-1) suggests antagonistic roles in Schwann cells. J. Neurosci. Res 50, 702–712 (1997).
Arroyo, E. J., Bermingham, J. R. Jr., Rosenfeld, M. G. & Scherer, S. S. Promyelinating Schwann cells express Tst-1/SCIP/Oct-6. J. Neurosci. 18, 7891–7902 (1998).
Sauka-Spengler, T. & Bronner-Fraser, M. A gene regulatory network orchestrates neural crest formation. Nat. Rev. Mol. Cell Biol. 9, 557–568 (2008).
Ma, K. H. & Svaren, J. Epigenomic reprogramming in peripheral nerve injury. Neural Regen. Res. 11, 1930–1931 (2016).
Arthur-Farraj, P. J. et al. Changes in the coding and non-coding transcriptome and DNA methylome that define the Schwann cell repair phenotype after nerve injury. Cell Rep. 20, 2719–2734 (2017).
Kim, Y. et al. The MMP-9/TIMP-1 axis controls the status of differentiation and function of myelin-forming Schwann cells in nerve regeneration. PLoS ONE 7, e33664 (2012).
Brushart, T. M. et al. Schwann cell phenotype is regulated by axon modality and central-peripheral location, and persists in vitro. Exp. Neurol. 247, 272–281 (2013).
Lin, H. P., Oksuz, I., Hurley, E., Wrabetz, L. & Awatramani, R. Microprocessor complex subunit DiGeorge syndrome critical region gene 8 (Dgcr8) is required for schwann cell myelination and myelin maintenance. J. Biol. Chem. 290, 24294–24307 (2015).
Pereira, J. A. et al. Integrin-linked kinase is required for radial sorting of axons and Schwann cell remyelination in the peripheral nervous system. J. Cell Biol. 185, 147–161 (2009).
Eshed-Eisenbach, Y., Gordon, A., Sukhanov, N. & Peles, E. Specific inhibition of secreted NRG1 types I-II by heparin enhances Schwann Cell myelination. Glia 64, 1227–1234 (2016).
Heraud-Farlow, J. E. et al. Protein recoding by ADAR1-mediated RNA editing is not essential for normal development and homeostasis. Genome Biol. 18, 166 (2017).
Bremer, J. et al. Ablation of Dicer from murine Schwann cells increases their proliferation while blocking myelination. PLoS ONE 5, e12450 (2010).
Topilko, P. et al. Krox-20 controls myelination in the peripheral nervous system. Nature 371, 796–799 (1994).
Verrier, J. D., Semple-Rowland, S., Madorsky, I., Papin, J. E. & Notterpek, L. Reduction of Dicer impairs Schwann cell differentiation and myelination. J. Neurosci. Res. 88, 2558–2568 (2010).
Jessen, K. R., Mirsky, R. & Lloyd, A. C. Schwann cells: development and role in nerve repair. Cold Spring Harb. Perspect. Biol. 7, a020487 (2015).
Quintes, S. & Brinkmann, B. G. Transcriptional inhibition in Schwann cell development and nerve regeneration. Neural Regen. Res. 12, 1241–1246 (2017).
Gasque, P. & Jaffar-Bandjee, M. C. The immunology and inflammatory responses of human melanocytes in infectious diseases. J. Infect. 71, 413–421 (2015).
Masaki, T. et al. Reprogramming adult Schwann cells to stem cell-like cells by leprosy bacilli promotes dissemination of infection. Cell 152, 51–67 (2013).
Kondo, T. et al. Six novel mutations of the ADAR1 gene in patients with dyschromatosis symmetrica hereditaria: histological observation and comparison of genotypes and clinical phenotypes. J. Dermatol. 35, 395–406 (2008).
Oyama, M., Shimizu, H., Ohata, Y., Tajima, S. & Nishikawa, T. Dyschromatosis symmetrica hereditaria (reticulate acropigmentation of Dohi): report of a Japanese family with the condition and a literature review of 185 cases. Br. J. Dermatol. 140, 491–496 (1999).
Peng, A. C. Y., Chen, Y. A. & Chao, S. C. Dyschromatosis symmetrica hereditaria: a retrospective case series and literature review. Dermatol. Sin. 31, 19–24 (2013).
Zhou, Q. et al. Two novel ADAR1 gene mutations in two patients with dyschromatosis symmetrical hereditaria from birth. Mol. Med. Rep. 15, 3715–3718 (2017).
Nordlund, J. J. et al. Vitiligo in patients with metastatic melanoma: a good prognostic sign. J. Am. Acad. Dermatol. 9, 689–696 (1983).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Naro, C. et al. An ochestrated intron retention program in meiosis controls timely usage of transcripts during germ cell differentiation. Dev. Cell 41, 82–93 e84 (2017).
Noli, L., Capalbo, A., Ogilvie, C., Khalaf, Y. & Ilic, D. Discordant growth of monozygotic twins starts at the blastocyst stage: a case study. Stem Cell Rep. 5, 946–953 (2015).
Traunmuller, L., Gomez, A. M., Nguyen, T. M. & Scheiffele, P. Control of neuronal synapse specification by a highly dedicated alternative splicing program. Science 352, 982–986 (2016).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Xiong, H. et al. RED-ML: a novel, effective RNA editing detection method based on machine learning. Gigascience 6, 1–8 (2017).
Opdecamp, K. et al. Melanocyte development in vivo and in neural crest cell cultures: crucial dependence on the Mitf basic-helix-loop-helix-zipper transcription factor. Development 124, 2377–2386 (1997).
Acknowledgements
The authors would like to thank members of the IMRB and LEAT-Imagine platforms for animal husbandry, members of the histology platform of Broussais Hospital for technical help, Yuli Watanabe and Edouard Reyez Gomez for help at the beginning of the project, Benoit Funalot and Yanick Crow for advices, Olivier Ariste and Ariane Jolly for help in data analysis, Elisabeth Dupin Veronique Delmas and Lionel Larue for advices on melanocytes rescue experiments, Frederic Rieux-Laucat for advices on immune cells recruitment and Brahim Nait Oumesmar for CD45 and (F4/80+CD11b+CD68) antibodies. This work was supported by the Fondation ARC pour la recherche sur le cancer and AFM trampoline grant to N.B., Institut National Pour la Santé de la Recherche Médicale (INSERM), state funding from the Agence Nationale de la Recherche under the “Investissements d’avenir” program [ANR-10-IAHU-01] and the MSDAvenir fund [DEVO—DECODE project]. N.G. and A.K. both received a 3-year Ph.D. fellowship from ED-SVS402, L.Z. received a 3-year Ph.D. fellowship from ED-BioSPC. N.G. is a beneficiary of an Institut Imagine 4th-year Ph.D. fellowship (7 months) from Fondation Bettencourt Schueller and supported by Fondation des treilles.
Author information
Authors and Affiliations
Contributions
N.G., A.K. and N.B. conceived, designed and performed the experiments and analyzed data; N.G. and N.B. wrote the manuscript and prepared figures; N.B. supervised the study and obtained ARC and AFM funding; L.Z. performed validation of RNA-seq data by RT-qPCR and helped with rescue experiments as well as immune cells and systemic response experiments; L.R. prepared samples for electronic microscopy; S.M. performed axon diameter quantification; R.P.K. analyzed gut histological sections; M.P. prepared libraries and performed sequencing; P.D.L.G. performed bioinformatics analysis of RNA-seq data; J.M.V. prepared and analyzed electronic microscopy data; S.D. provided the HtPA-Cre line; V.P. discussed results; V.P., S.D., L.Z., R.P.K., J.A. made critical revision of the manuscript and all authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Communications thanks Lukas Sommer, Carl Walkley and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gacem, N., Kavo, A., Zerad, L. et al. ADAR1 mediated regulation of neural crest derived melanocytes and Schwann cell development. Nat Commun 11, 198 (2020). https://doi.org/10.1038/s41467-019-14090-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-019-14090-5
This article is cited by
-
Melanocytes in regenerative medicine applications and disease modeling
Journal of Translational Medicine (2024)
-
MAIT cell inhibition promotes liver fibrosis regression via macrophage phenotype reprogramming
Nature Communications (2023)
-
Multi-dimensional Insight into the Coexistence of Pathogenic Genes for ADAR1 and TSC2: Careful Consideration is Essential for Interpretation of ADAR1 Variants
Biochemical Genetics (2023)
-
Direct identification of A-to-I editing sites with nanopore native RNA sequencing
Nature Methods (2022)
-
Aicardi-Goutières syndrome-associated mutation at ADAR1 gene locus activates innate immune response in mouse brain
Journal of Neuroinflammation (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.