- 1 Marine Biological Laboratory, The Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Woods Hole, MA, USA
- 2 H. Steinitz Marine Biology Laboratory, Interuniversity Institute for Marine Sciences, Eilat, Israel
- 3 Institute of Marine Sciences, University of California Santa Cruz, Santa Cruz, CA, USA
Spatial patterns of marine Synechococcus diversity across ocean domains have been reported on extensively. However, much less is known of seasonal and multiannual patterns of change in Synechococcus community composition. Here we report on the genotypic diversity of Synechococcus populations in the Gulf of Aqaba, Northern Red Sea, over seven annual cycles of deep mixing and stabile stratification, using ntcA as a phylogenetic marker. Synechococcus clone libraries were dominated by clade II and XII genotypes and a total of eight different clades were identified. Inclusion of ntcA sequences from the Global Ocean Sampling database in our analyses identified members of clade XII from beyond the Gulf of Aqaba, extending its known distribution. Most of the Synechococcus diversity was attributed to members of clade II during the spring bloom, while clade III contributed significantly to diversity during summer stratification. Clade XII diversity was most prevalent in fall and winter. Clade abundances were estimated from pyrosequencing of the V6 hypervariable region of 16S rRNA. Members of clade II dominated Synechococcus communities throughout the year, whereas the less frequent genotypes showed a pattern of seasonal succession. Based on the prevailing nutritional conditions we observed that clade I members thrive at higher nutrient concentrations during winter mixing. Clades V, VI and X became apparent during the transition periods between mixing and stratification. Clade III became prominent during sumeer stratification. We propose that members of clades V, VI, and X, and clade III are Synechococcus ecotypes that are adapted to intermediate and low nutrient levels respectively. This is the first time that molecular analyses have correlated population dynamics of Synechococcus genotypes with temporal fluctuations in nutrient regimes. Since these Synechococcus genotypes are routinely observed in the Gulf of Aqaba we suggest that seasonal fluctuations in nutrient levels create temporal niches that sustain their coexistence.
Introduction
The abundant marine picocyanobacteria Synechococcus and Prochlorococcus display high genotypic diversity, both among culture isolates and in natural samples (Ferris and Palenik, 1998; Moore et al., 1998; Rocap et al., 2002; Fuller et al., 2003; Penno et al., 2006). Much is known about the spatial and temporal distributions of Prochlorococcus in its habitat between latitudes 40′N and 40′S (Moore et al., 1995; Johnson et al., 2006). Although Prochlorococcus is often more abundant than Synechococcus, its numbers decline sharply in upwelling systems and basins with deep convective mixing in winter (Lindell and Post, 1995; Partensky et al., 1999; Durand et al., 2001). Prochlorococcus populations span the photic zone and are found down to 200 m depth (Olson et al., 1990; Veldhuis and Kraay, 1990; Lindell and Post, 1995; Partensky et al., 1999). The Prochlorococcus lineage contains six well-defined clades (Moore et al., 1998; West and Scanlan, 1999; Rocap et al., 2002) with fully sequenced genomes of representative strains (Kettler et al., 2007) but novel clades have been reported recently (Martiny et al., 2008; Kamennaya and Post, in preparation). Genotypes of the high light (HL) adapted clades I and II and low light (LL) adapted clades I, II, III, and IV were proposed to represent ecotypes as their distributions often reflect prevalence for certain ocean niches (Johnson et al., 2006; Zinser et al., 2006; Garczarek et al., 2007).
Synechococcus populations extend into more temperate waters and they are abundant in the surface mixed layer (approx. 0–50 m) of stratified water bodies (Lindell and Post, 1995; Partensky et al., 1999). Synechococcus isolates are surprisingly diverse and their phylogenies have been studied using 16S rRNA, ITS, rpoC, cpeAB, narB, and ntcA as molecular markers (Toledo and Palenik, 1997; Rocap et al., 2002; Fuller et al., 2003; Steglich et al., 2003; Ahlgren and Rocap, 2006). Synechococcus populations are genetically more diverse than those of Prochlorococcus and at least 16 clades (I–XVI) have been distinguished (Fuller et al., 2003; Ahlgren and Rocap, 2006; Penno et al., 2006). Both 16S and concatenated core genome phylogenies have identified two main Synechococcus clusters, 5.1a and 5.1b (Figure 1), representing open ocean and coastal Synechococcus types respectively (Dufresne et al., 2008). These are rather broad definitions when taking into account that their accessory genomes indicate pronounced differences in their ability to respond to environmental cues (Scanlan et al., 2009). Quantitative approaches employing clade-specific oligonucleotides have shown that clade II Synechococcus is the dominant genotype in the northern Red Sea and the Arabian Sea (Fuller et al., 2005, 2006a) and among culture isolates from both those environments (Fuller et al., 2003). The first hints that Synechococcus clades correspond to ecotypes were observed in the Arabian Sea with clade II genotypes dominating in central and northern parts, and clade III more abundant at more southern sites along the transect (Fuller et al., 2006a). Genotypes belonging to either clade V, VI, or VII were found to dominate Synechococcus populations in upwelling regions (Fuller et al., 2006a), suggesting that their distribution is a reflection of high nutrient availability at such locations. Subsequently, dot blot and FISH analyses along a transect in the Atlantic Ocean indicated that distributions of Synechococcus clades are remarkably similar for corresponding regions in the Northern and Southern hemispheres (Zwirglmaier et al., 2007). A complementary study identified the global distribution patterns for Synechococcus clades I–VII (Zwirglmaier et al., 2008). Both studies concluded that Synechococcus communities do not change significantly with depth – as seen in Prochlorococcus – but rather on a horizontal scale. The highest abundances of clades I and IV are in coastal and temperate mesotrophic waters (above 30′N or below 30′S) whereas clade II dominates oligotrophic (sub)tropical offshore waters and clade III the oligotrophic open ocean waters. Clade V, VI, and VII genotypes (studied as single group in dot blot hybridizations) were detected in low abundance and their functional diversity is still poorly understood (Zwirglmaier et al., 2008).
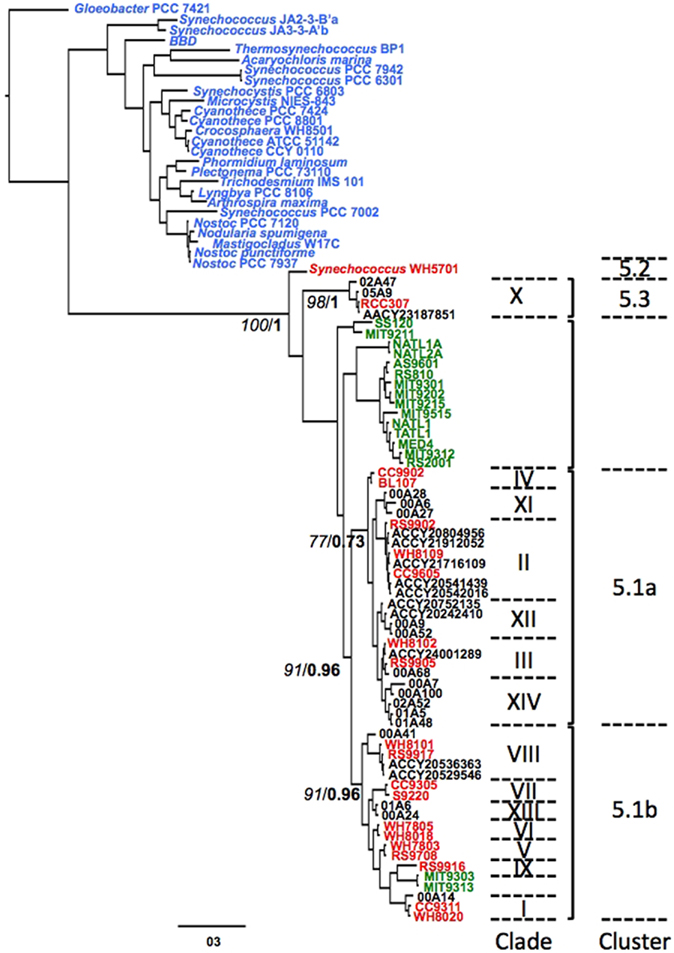
Figure 1. A gene tree of full-length ntcA sequences retrieved from GenBank, including representative strains of all known Synechococcus (red, clades marked with Roman numbers) and Prochlorococcus clusters (green). All other cyanobacteria ntcA are indicated in blue. Numbers at nodes denote bootstrap support (RAxML, italics) and posterior probability (MrBayes, bold). Environmental sequences from the Gulf of Aqaba (format xxAyy, in which xx is year, A denotes station A in open waters and yy denotes clone number) were used to represent clusters for which no culture isolates are available. Sequences identified by their ACCY prefix were retrieved from the Global Ocean Survey (GOS) database using Synechococcus NtcA sequences as query in blastn searches.
Current concepts of picocyanobacterial distribution are largely shaped by the spatial dimensions of Synechococcus niches; temporal aspects of their distributions have received little attention. Succession among Synechococcus genotypes was described for coastal waters off California where for three consecutive years members of clades II and III were prominent in the months leading to the Synechococcus spring bloom while clades I and IV dominated the bloom itself (Tai and Palenik, 2009). The oligotrophic Gulf of Aqaba, Northern Red Sea is subject to dramatic annual alternation between deep winter mixing and summer stratification (Wolf-Vecht et al., 1992; Genin et al., 1995). Synechococcus and Prochlorococcus numbers change over 3–4 orders of magnitude during the seasonal cycle (Lindell and Post, 1995), but little is known of how this reflects on the genotypic composition of picocyanobacterial communities. We have previously employed the nitrogen regulatory gene ntcA as a molecular marker for phylogenetic studies of cyanobacteria (Lindell et al., 2005; Penno et al., 2006). This approach specifically targets cyanobacteria, distinguishes individual clades with a high resolution and led to the identification of 4 novel Synechococcus clades (Penno et al., 2006), in addition to the 10 known clades (Rocap et al., 2002; Fuller et al., 2003). Here we report on a seasonal succession among Synechococcus genotypes following a deep mixing event in the Gulf of Aqaba. Based on correlations between hydrographic/nutritional conditions and genotype abundance we assign trophic ecotypes to a number of Synechococcus clades.
Materials and methods
Sampling Procedures
Seawater was collected monthly at discrete depths along a 730-m deep water column at station A (29°28′N, 34°55′E), a sampling site in the open waters of the Gulf of Aqaba, northern Red Sea. Sampling was mostly performed in conjunction with activities for larger monitoring programs like the “Gulf of Aqaba Peace Park” Project (1999–2002) and the National Monitoring Program (NMP, Israel; 2003–2006) for the entire research period from March 2000 to May 2006. A CTD-Rosette sampler equipped with 12 L Teflon-coated GO-FLO bottles (General Oceanics, Inc., Miami, USA) was used to obtain water samples while simultaneously recording profiles of chlorophyll a, salinity and temperature with depth (Seacat SBE19, Seabird electronics, Inc.). Light intensity and chlorophyll a fluorescence profiles were obtained with a LI-COR light sensor (LI-192SA) and a Seapoint Sensors fluorometer mounted on the CTD-Rosette. For DNA analyses 5–20 L samples were passed over a 20-μm mesh and stored at 4°C in darkness. Samples were filtered within 24 h onto 0.2 (16S pyrosequencing) or 0.45 μm (ntcA PCR) phenol-soluble polysulfone filters (Gelman Supor-450, Ø 47 mm) under gentle vacuum (<15 mm Hg). The filters were then placed in 4 mL DNA extraction buffer (750 mM sucrose, 400 mM NaCl, 20 mM EDTA, 50 mM Tris–HCl, pH 9), quickly frozen in LN2 and stored at −20°C. For cell counts duplicate 1.5 mL samples were fixed with 0.2% paraformaldehyde (pH 8.0) for 20 min at room temperature in the dark, frozen in LN2 and stored at −80°C.
Determination of Nutrients and Bacterial Cell Numbers
Concentrations of nitrate, nitrite, and phosphate in seawater samples were determined using a flow injection autoanalyzer (FIA, Lachat Instrument Model Quickchem 8000) and are available from the NMP (www.iui-eilat.ac.il/NMP). Ammonium concentrations were measured using a fluorometric method using a modified protocol based on Holmes et al. (1999). Synechococcus cell numbers were determined with a FACScan flow cytometer (Beckton Dickenson) with 15 mW neon-argon laser excitation at 488 nm (Marie et al., 1999).
DNA Extraction and PCR Amplification
Samples were thawed on ice and DNA was extracted using phenol–chloroform followed by isopropanol–ammonium acetate precipitation according to Penno et al. (2006). Up to 50 ng of genomic DNA was used in PCR amplification of ntcA as described in Lindell et al. (1998) using 20 μM of the cyanobacteria-specific, degenerate primer pairs 1F and 4R or 1AF and 4AR in the ratio of 1:1 or 3:1, respectively (see Lindell et al., 1998). These primers anneal to conserved regions of the ntcA gene and amplify a 449-bp fragment. PCR reactions were performed using a PTC-200 thermal cycler (MJ Research, Inc.) with an initial denaturation of 4 min at 94°C followed by 35–40 cycles of 1 min at 94°C, 1 min at 50–55°C, 1 min at 72°C, and finally a 5-min extension period at 72°C.
ntcA Clone Libraries and Sequence Analyses
For ntcA clone libraries agarose gel extraction (QIAquick gel extraction kit, Qiagen) was used to purify the 449-bp PCR products, which were cloned into a pGEM-T vector (Promega). Clones were transformed into competent E. coli strain DH5α cells following standard procedures and plated on ampicillin, X-Gal/IPTG selective LB plates for blue/white selection. The ntcA clone libraries were screened for Synechococcus sequences by PstI digestion (1 h at 37°C) of purified plasmids (Miniprep, Qiagen; Penno et al., 2006). In some instances clones were inspected by nested PCR using Synechococcus specific primers G15/16F and SR50 (Lindell and Post, 2001). At least 200 ng purified plasmid DNA (Miniprep, Qiagen) or 50 ng purified PCR product was used for sequence reactions using the BigDye Terminator cycle sequencing chemistry (Applied Biosystems) and analyzed on an ABI 3700 instrument at the Genome Services Center at the Hebrew University Jerusalem. Resulting sequences were compared to NCBI GenBank using BLASTN or BLASTX. The Synechococcus ntcA sequences were deposited in NCBI GenBank with accession numbers DQ204774–DQ204834, DQ204836, DQ204838, DQ204839, DQ204841–DQ204868.
Phylogenetic Analyses
Sequences were aligned using ClustalW and ClustalW2 (EMBL-EBI, BioEdit version 5.0.9, Hall, 1999) and MUSCLE (Edgar, 2004); the Bayesian tree was generated with MrBayes 3.1 (Huelsenbeck and Ronquist, 2001) and the maximum likelihood tree with RAxML 7.2.8 (Stamatakis, 2006) using the general time reversible model and rate variation modeled using a gamma distribution (“nst = 6 rates = gamma for MrBayes”; “-m GTRGAMMA” for RaxML); model parameters for codon third positions were estimated independently of codon first and second positions (“unlink revmat statefreq shape” for MrBAyes; “-q” for RAxML). For MrBayes, two independent runs of 4 chains each were run for 2 million generations and sampled every 100 generations; comparison of the parameter estimates from the two runs indicated convergence (Gelman and Rubin, 1992). The first hundred thousand trees were discarded as burn in before generating the consensus tree. For RAxML, the best tree was chosen from 100 iterations of both maximum parsimony and random starting trees, using an empirically determined initial rearrangement setting of 10; the results of 1000 bootstrap iterations were superimposed on the best tree.
Pyrosequencing
Environmental DNA (30 ng) from monthly samples taken during the 2006–2007 annual cycle were templates for PCR with eubacterial primers that target the flanking regions of the V6-hypervariable region of the small subunit 16S rRNA; the resulting amplicons were sequenced on a Roche GS FLX as previously described (Huber et al., 2007). After removing low-quality reads (Huse et al., 2007), each sequence was assigned taxonomy based on comparison to the SILVA reference database of bacterial 16S sequences (Huse et al., 2008). Sequences with their abundances and taxonomic identification are available for each sample at http://vamps.mbl.edu under the project “ICM_GOA.”
Results
ntcA Phylogeny
We retrieved over 60 full-length ntcA accessions from cyanobacterial strains from NCBI GenBank to construct Bayesian and maximum likelihood gene trees (Figure 1). The two methods yielded near-identical tree topologies, which served as the backbone structure for clade assignment of our environmental sequences (see below). Marine Synechococcus and Prochlorococcus formed a monophyletic lineage with the Synechococcus euryhaline strain WH5701 (sub-cluster 5.2) and marine strain RCC307 (sub-cluster 5.3) basal to other picocyanobacteria. Most Prochlorococcus ntcA sequences formed a lineage distinct from Synechococcus sub-clusters although LL adapted strains MIT9313 and MIT9303 (Prochlorococcus clade LL IV) were found in Synechococcus sub-cluster 5.1b, a recurring observation in phylogenies of both ntcA (Lindell et al., 2005; Penno et al., 2006) and the urea transport gene urtA (Kamennaya et al., 2008).
Synechococcus clades V, VI, VII, VIII, and IX, and clades II, III, and IV were members of sub-clusters 5.1a and 5.1b, respectively, consistent with trees based on 16S rRNA and on concatenated alignments of >1000 genes in the Synechococcus core genome (Dufresne et al., 2008). We included novel Synechococcus clades XI, XII, XIII, and XIV – identified in ntcA phylogenies of environmental sequences from the Gulf of Aqaba (Penno et al., 2006) – in our analysis and representative sequences for each clade are presented in Figure 1. We found that clades XI, XII, and XIV are members of sub-cluster 5.1a and thus are likely open ocean ecotypes, whereas clade XIII is a member of sub-cluster 5.1b and is thus likely a coastal ecotypes (Dufresne et al., 2008). Lastly, we searched the Global Ocean Sampling (GOS) database for ntcA sequences using BLASTN with environmental Gulf of Aqaba sequences from clades XI, XII, XIII, and XIV as queries. We found 11 (nearly) full-length ntcA sequences; we could assign eight sequences to clades II, III, or VIII of sub-clusters 5.1a or b and one clustered to sub-cluster 5.3 (Figure 1). The remaining two GOS sequences were loosely affiliated with clade XII and shared 81–83% identity with clade XII clones from the Gulf of Aqaba.
Environmental Synechococcus ntcA Sequences
Following the cyanobacterial diversity studies at sampling station A in the Gulf of Aqaba in 1998–2000 (Fuller et al., 2003, 2005; Lindell et al., 2005; Penno et al., 2006), we re-sampled this site over six consecutive years with a focus on succession patterns in surface waters (0–20 m) following the annual Synechococcus spring bloom. We obtained a total of 354 unique sequences of Synechococcus ntcA, primarily from samples collected at 20 m depth (Table 1). In a previous study we showed that 16S and ntcA phylogenies yield identical branching patterns and resolved ntcA genotype clusters (clades) at a threshold of 85% nucleotide identity (Penno et al., 2006). Using this criterion, all but two environmental sequences were assigned as members of known Synechococcus clades (Table 1). We identified eight different Synechococcus clades, four of which (clades I, II, III and sub-cluster 5.3) are represented by culture strains in the databases. Four additional clades observed in the Gulf of Aqaba (XI, XII, XIII, and XIV) still lack culture representatives. Sequences 00A100 and 02A47 are most closely related to sub-cluster 5.3 but shared only 78–79% identity with members of that group. The rarefaction analysis indicated that sampling of Synechococcus diversity was nearly saturated at the clade level (Figure 2). The bulk of environmental sequences (>96%) obtained during this 7 year study belonged to Synechococcus subgroup 5.1a. Most abundant were members of clade II (57%) and clade XII (22%). Members of sub-clusters 5.1b and 5.3 (clade X) were detected but at much lower frequencies (<4%).
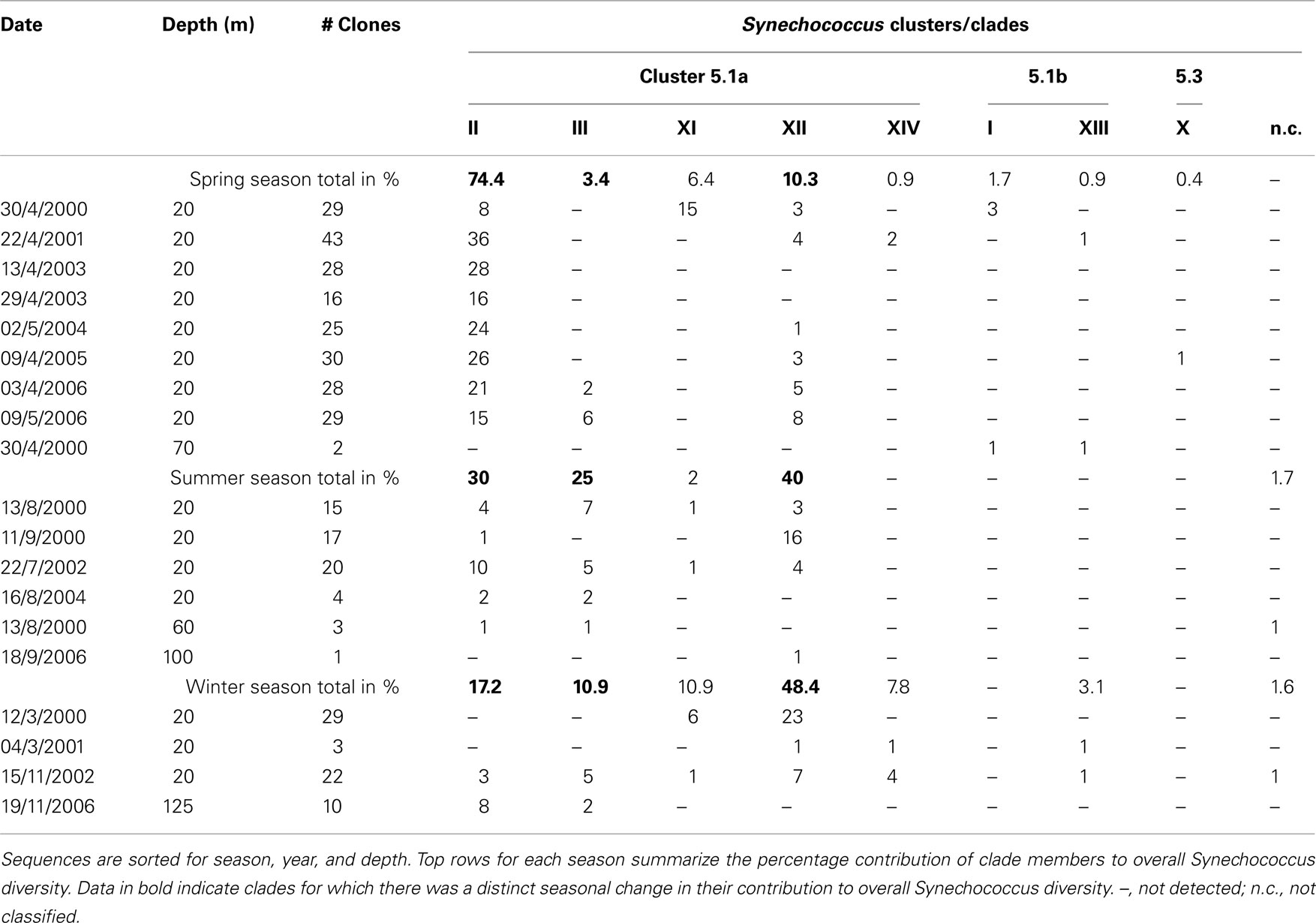
Table 1. Overview of all unique ntcA sequences and their affiliation with different Synechococcus clades.
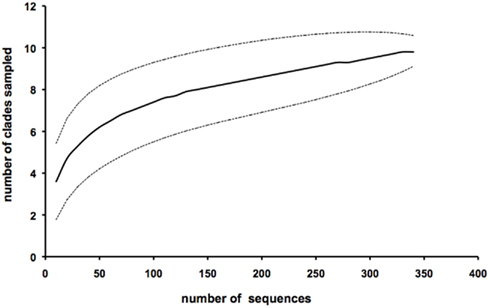
Figure 2. Cluster analysis of all 354 environmental ntcA sequences from the Gulf of Aqaba using Analytic Rarefaction 1.3 Software (Holland, 2003, http://www.uga.edu/strata/software/). The dotted lines show the upper and lower 95% confidence limits. Sequences were analyzed as a 15% threshold in accordance with the definition of genotype clusters in Penno et al. (2006).
Temporal Distribution of Synechococcus Genotypes
Members of the two main Synechococcus sub-clusters, 5.1a and 5.1b, have been distinguished by their numerical dominance in different habitats, dominating in open ocean (specialist) and coastal (opportunist) environments, respectively (Dufresne et al., 2008). In this study we investigated whether members of different clades may represent different ecotypes adapted to specific light-nutrient combinations. The Gulf of Aqaba is subject to an annual deep mixing event in winter, accompanied by nutrient injection into surface layers. Deep mixing starts in October and reaches maximal depths in late February/March (Figure 3A). Nutrients, such as nitrate, nitrite, ammonium (Figure 3B), and phosphate (not shown) at 20 m depth reach maximal concentrations during the same period, while the increase in chlorophyll a concentrations and Synechococcus cell numbers follow with a 2 to 4-week delay (Figure 3C). Nitrate and phosphate concentrations at 20 m depth showed a positive correlation with mixing depth (Figure 4) with R2 values of 0.650 and 0.576, respectively. Nitrate and phosphate were present in the mixed layer at an approximate ratio of 9:1. Substantial nitrite concentrations in the mixed layer at these times support an inorganic N:P ratio approaching 16:1 (Mackey et al., Submitted). Deep mixing events were followed by distinct Synechococcus spring blooms (Lindell and Post, 1995) after which their abundance steadily dropped in the nutrient-deplete, stratified summer waters (Figure 3C).
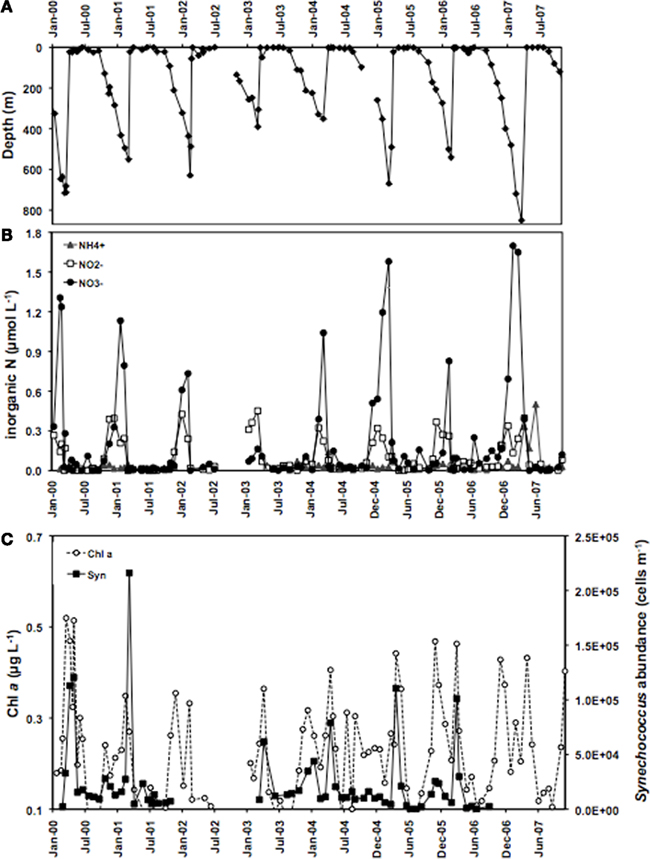
Figure 3. Overview of (A) mixing depth, (B) nitrite, nitrate, and ammonium concentrations at 20 m depth and (C) chlorophyll a concentrations and Synechococcus cell numbers at 20 m depth at sampling station A in open waters of the Gulf of Aqaba between 2000 and 2006.
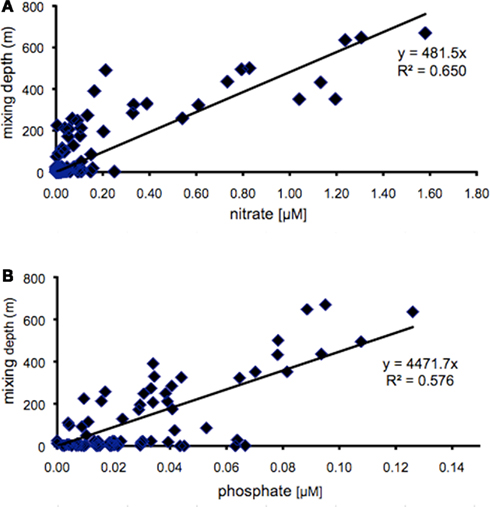
Figure 4. Surface mixed layer concentrations (sampled at 20 m) of nitrate (A) and phosphate (B) as function of mixing depth at sampling station A between 2000 and 2006. Line indicates the curve resulting from linear regression analysis.
Previously we found that the most abundant Synechococcus clade in the Gulf of Aqaba was clade II (Fuller et al., 2003). In the present study, spanning 7 years, we found annual trends of change in the genotypic composition of Synechococcus populations, in good correlation with macronutrient availability. Table 1 summarizes the total clone numbers for different seasons and their assignment to different clades. Members of novel clades XI–XIV were mostly observed in winter and spring of years with truly deep mixing (>500 m) and high nutrient levels (Table 2). Synechococcus diversity in spring (immediately following the deep mixing event) was mostly contributed by members of clade II. In years when mixing is only moderate (<400 m in 2003 and 2004) this dominance is encompassing, while additional clades contributed significantly to the diversity (14–72%) in other years. This suggests that the extent (depth and duration) of mixing events is a significant factor in shaping the Synechococcus diversity in the Gulf of Aqaba. The number of unique clade II sequences averaged 23 ± 8 in spring; this number fell below 10 during summer and winter. Clade III members were more prominent during late spring and summer.
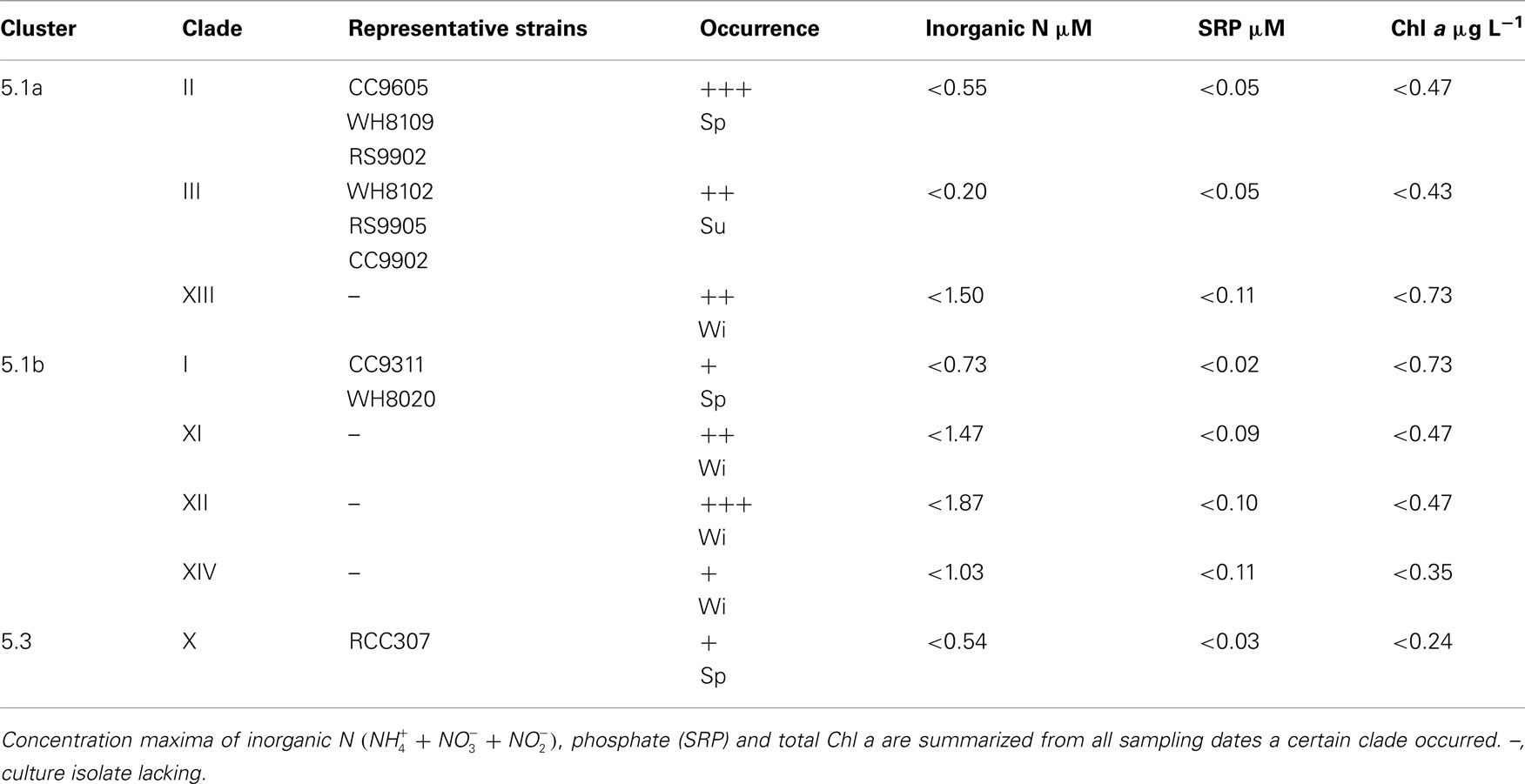
Table 2. Representation of Synechococcus genotypes in ntcA clone libraries with + low, ++ intermediate, and +++ high frequency occurrence in their season of prevalence (Sp, spring; Su, summer; Wi, winter).
We summed the contributions of the individual clades to arrive at a trend in seasonal change of Synechococcus diversity (Table 1). More than 95% of the diversity was contributed by clades within sub-cluster 5.1a. Of these, clade II made up 74.4% of the Synechococcus diversity during the spring bloom, but fewer clade members were identified in other seasons and their contribution to Synechococcus diversity fell to 17.2% in winter. Clade III genotypes were observed from April through November, but they were lacking from winter (December–March) samples (Table 1). This clade contributed 25% of the diversity in summer. The last clade that featured dominantly in Synechococcus diversity was clade XII. Although it contributed only 10% during the spring bloom this number rose to 40–48% in summer and winter. Together these findings indicate that Synechococcus diversity in the Gulf of Aqaba is subject to a seasonal pattern and possibly that members of clade II, III, and XII occupy temporal niches that select for the ecotypes contained in these clusters. We studied quantitative changes in Synechococcus populations as part of a deep sequencing effort of microbial community structures over the 2006–2007 annual cycle.
Pyrosequencing of the V6 Hypervariable Region of 16S rRNA
In order to confirm some of the trends evident in the ntcA data we quantified the contribution of various Synechococcus clades over the 2006–2007 annual cycle using massively parallel tag sequencing analysis of microbial community samples (Sogin et al., 2006). A total of 516,181 sequences spanning the V6 hypervariable region of the bacterial 16S gene were obtained from monthly samples (5–20 m depth). Of these, 45,844 were identified (Huse et al., 2008) as cyanobacterial with 28,798 tags assigned to Prochlorococcus and 17,046 to Synechococcus. To improve taxonomic resolution we inspected the V6 region of representative Synechococcus strains for which genome sequences are available. Genome analysis classified these strains as representing open ocean or coastal habitats (Dufresne et al., 2008). An alignment showed nucleotide differences at multiple positions among the majority of these strains in the V6 region (Figure 5). Seven Synechococcus groups can be distinguished based on two or more nucleotide differences from the other sequences (Figure 6): clade I, clade II, clades III/IV (the V6 region is identical in cultured isolates of these two clades), clade V/VI (differ by a single nucleotide), clade X and the estuarine strain WH5701. Comparing the Synechococcus V6 tags against the NCBI refseq_genomic database using blastn resolved five different Synechococcus groups by sequence similarity: clades I, II, III/IV, V/VI, and X (Figure 6). As with the ntcA clone libraries, none of the V6 sequences could be assigned to clade IX or clade VIII, although both clades are defined based on isolates from the Gulf of Aqaba. Total Synechococcus V6 tag abundances closely mimicked actual Synechococcus population dynamics over the seasonal cycle (Lindell and Post, 1995; Penno et al., 2006). Consistent with the ntcA results, clade II-like V6 tags dominated the Synechococcus population throughout the 2006–2007 period. Clade III/IV tags were more pronounced during the summer months (12 ± 4% of total as compared to 3 ± 1% in winter), though at abundances an order of a magnitude lower than clade II-like V6 tags (Figure 7). Our results indicated that clades with low tag abundances engaged in a seasonal succession. Clade I-like V6 tags were observed only during the winter mixing, together with clade V and clade X-like tags. The latter two groups preceded the rise of clade I-like tags during the transition phase from nutrient-deplete to nutrient-rich waters. They also persisted through the transition phase from nutrient-rich to nutrient-deplete waters in the stably stratified Gulf of Aqaba that eventually lead to a more prominent presence of clade III/IV-like tags.

Figure 5. Alignment of the 60 nucleotide V6 hypervariable region (position 904–963 in strain CC9902) of the small subunit (16S) ribosomal rRNA gene in 11 marine/estuarine Synechococcus genomes. Nucleotide identities are indicated with “*”.
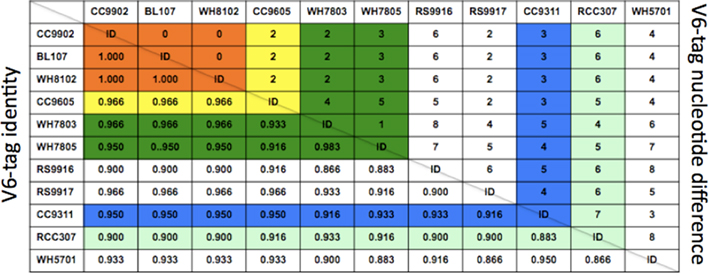
Figure 6. Identity matrix (bottom half) and nucleotide difference of 60 nucleotide V6 hypervariable region of the small subunit (16S) ribosomal rRNA gene in 11 marine/estuarine Synechococcus genomes. Strain CC9902 was taken as the basis for nucleotide identity calculations. ID, identical.
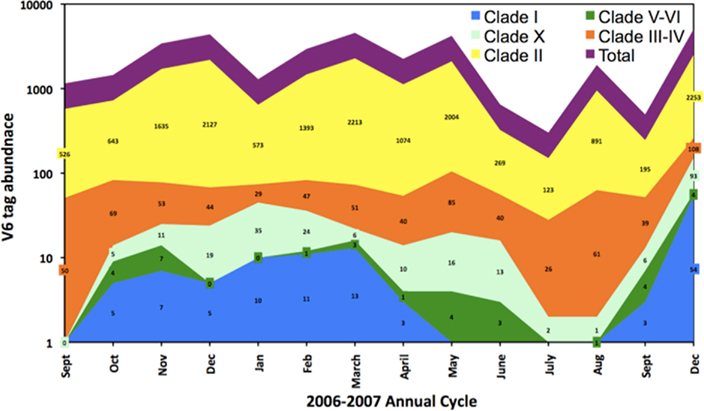
Figure 7. V6 tag abundance of Synechococcus ecotypes in surface layer samples (20 m) in the Gulf of Aqaba along a seasonal cycle in 2006–2007. Data labels indicate the actual V6 tag abundance of each clade for individual samples and the total V6 tag abundance (purple) is the sum of these numbers. Color codes for the various clades are identical to those of their “type” strains for which V6 sequence identities were inspected (see Figure 6).
Discussion
In this study we followed the seasonal change among Synechococcus genotypes over a 7-year period in the Gulf of Aqaba. We observed a high diversity involving members of eight different clades. We show that the genotypes contained in the various clades likely represent ecotypes, each selected for under a different set of environmental conditions. Three low abundance ecotypes engage in a seasonal succession following annual deep mixing events. The seasonal succession of ecotypes underscores the year-round dominance of Synechococcus at 104–105 cells mL−1 in the surface waters of the Gulf of Aqaba. This dominance contrasts with the population dynamics of Prochlorococcus that fluctuate between 105 cells mL−1 in summer and 102 cells mL−1 in winter in the same waters (Lindell and Post, 1995).
Synechococcus Diversity
Synechococcus ntcA sequences from the Gulf of Aqaba were clustered at the 15% difference level, forming eight clusters, each corresponding to a previously recognized clade. Rarefaction analysis suggests that the diversity of 15% clusters was essentially completely sampled, although additional clades (V, VII, VIII, and IX) have been reported from the Gulf in previous studies using ntcA (Fuller et al., 2003; Lindell et al., 2005). These clades were probably present as low abundance genotypes during 2000 to 2006 and therefore not detected with our PCR protocols. High throughput sequencing of 16S V6 tags yielded low abundant sequences similar to RCC307 (clade X) and WH7803 (clade V) genotypes, but not clades VII, VIII, and IX. Altogether the Synechococcus diversity in the Gulf of Aqaba is among the highest reported with members of 12–13 clades (out of a total of 16) identified. In comparison, studies of the California Current (Tai and Palenik, 2009), Sargasso Sea (Ahlgren and Rocap, 2006) and Arabian Sea (Fuller et al., 2006b) identified 6–8 Synechococcus clades. The diversity in the Gulf of Aqaba is in part contributed by novel clades XI through XIV. Members of these clades are frequently observed in the Gulf, but have not been reported from other locations (with the exception of clade XII, for which we identified sequences entries in the GOS database). Clades XI, XIII, and XIV have to date been reported only from the Gulf of Aqaba. Massively parallel V6 tag sequencing resolved fewer clades than the ntcA amplicons libraries. It was previously shown that clade IV strains BL107 and CC9902 have identical 16S sequences (Dufresne et al., 2008; Scanlan et al., 2009). Here we show that clades III and IV cannot be distinguished by their V6 sequences, nor can clades V and VI. Our ntcA phylogeny (Figure 1) indicates that novel clade XIII is closely related to clades V and VI, while clades XII and XIV are closely related to clade II. Therefore, tags identified in our V6 database as clade II and clades V + VI may actually include members of these novel clades. Since we showed here that, e.g., clade XII contributes to seasonal dynamics, it is clear that seasonal patterns cannot be fully resolved from V6 tag analyses.
Synechococcus Distributions
Previous publications have correlated Synechococcus abundance and diversity to its occurrence in different habitats. Highest abundances are found in upwelling and deep mixing regimes as well as over continental shelves (Olson et al., 1990; Lindell and Post, 1995; Moore et al., 1995; Durand et al., 2001; Zwirglmaier et al., 2007), which are nutrient-rich environments in comparison to the oligotrophic ocean gyres dominated by Prochlorococcus (Partensky et al., 1999; Johnson et al., 2006). Comparative genome studies divided marine Synechococcus into specialist or open ocean types versus an opportunist or coastal types (Dufresne et al., 2008; Scanlan et al., 2009). In contrast, field studies defined geographical domains for different Synechococcus genotypes, correlating their relative abundance to temperature. Members of clades II and III are more dominant in warm (sub)tropical waters while clades I and IV dominate in polar and temperate waters, respectively (Zwirglmaier et al., 2007). At a global scale genotypes V and VII are widely distributed albeit at moderate to low abundance (Zwirglmaier et al., 2007, 2008). However, it is not clear how well these habitats are defined. For example, clade I Synechococcus has been observed in the warm open waters of the Arabian Sea (Fuller et al., 2006b) and Red Sea (Penno et al., 2006); it dominates Synechococcus communities in temperate waters of Monterey Bay (Post and Zehr, unpublished results) and the New England Shelf (Post and Hunter-Cevera, unpublished results). Clade I Synechococcus was a feature in deeply mixed winter waters of the Gulf of Aqaba (Table 1; Figure 7). In our analyses, members of clades II, III, XI, and XIII dominated the community with lesser contributions of clades I and XIV. Typical coastal Synechococcus clades V–IX were either absent or a minor component of our extensive datasets for ntcA and V6. Despite its proximity to land, the Gulf of Aqaba thus has the characteristics of open ocean waters. Although this characterization has been made previously based on hydrographic arguments (Lindell and Post, 1995) this is the first time these water masses are classified through diversity analyses. We note that the distinction of open ocean and coastal ecotypes refers to their geographical distribution and not to their ecological physiology. Changes in diversity and abundance of the various clades in the Gulf of Aqaba (Penno et al., 2006) correlated with changes in the N status of the Synechococcus community in different seasons (Lindell and Post, 2001; Lindell et al., 2005). We suggest that clades contained in sub-clusters 5.1a and 5.1b represent ecotypes that differ in their nutrient requirements; clades contained in sub-cluster 5.1a are more typical of nutrient poor waters, while those of sub-cluster 5.1b are found in waters with higher nutrient availability. Seasonal fluctuations in nutrient availability in the Gulf of Aqaba create temporal niches that sustain the coexistence of clade representatives of both sub-clusters in the same water body. Temporal variation of Synechococcus clades has also been reported for a Pacific coastal site (Tai and Palenik, 2009). The variation at this location involved clade I and IV genotypes mostly, with minor contributions of clades II and III (Tai and Palenik, 2009). Although clade II-like V6 tags were dominant in the Gulf of Aqaba throughout the year, a number of Synechococcus clades participated in a seasonal succession, following the annual deep mixing event. Analysis of our V6 data indicated that this succession involves three major groups: clade I-like Synechococcus during deep winter mixing, clade III-like during summer stratification and clade V + X-like during the transition periods. Naturally, longer, smaller ntcA amplicon libraries yield a higher phylogenetic (Penno et al., 2006) but lower quantitative resolution than shorter, larger 16S rRNA libraries. Notwithstanding the fact that seasonal patterns of ntcA clades were more complex, a pattern consistent with that of the V6 tags emerged (Figure 5): a dominance of clade II and a presence of clade I during the spring bloom, while during summer clade III becomes more dominant. Interestingly, novel clade XII is also a major contributor to this seasonal succession and it becomes prominent in summer along with its close relative clade III (see Figure 1; Table 1). Since the Gulf of Aqaba has relatively small variations in light (1700–2000 μmol quanta m−2 s−1) and temperature (21–26°C) over the annual cycle, successions among Synechococcus groups may thus be more closely correlated with nutrient availability (Lindell and Post, 1995; Post et al., 2002). We propose that the low abundance Synechococcus clades represent ecotypes that track seasonal trophic gradients in the Gulf of Aqaba and engage in a seasonal succession.
Synechococcus Ecotype Succession
In accordance with the nutrient regimes at diverse locations, clade I is typical of eutrophic waters in more temperate climate zones and clades III–IV are typical of oligotrophic ocean waters (Fuller et al., 2006b; Zwirglmaier et al., 2007, 2008; Tai and Palenik, 2009). Clades V–VII are commonly found over continental shelves, environments with dynamic changes in nutrient and light supply (Fuller et al., 2006b; Zwirglmaier et al., 2007, 2008; Tai and Palenik, 2009), often mesotrophic conditions. Clade II is found in most (sub)tropical marine waters across a broad range of nutrient concentrations (Fuller et al., 2003, 2005, 2006b; Lindell et al., 2005; Penno et al., 2006; Zwirglmaier et al., 2007, 2008; Tai and Palenik, 2009). Nevertheless these ecotypes are all present in open waters of the Gulf of Aqaba. With inorganic N and P concentrations of <3 and <0.2 μM, chlorophyll a at <1 μg L−1 and 1% light depth at 80–90 m (Lindell and Post, 1995; Lindell et al., 2005; Penno et al., 2006) the Gulf is oligotrophic by any standard. Still, hydrographic conditions cause distinct nutrient regimes that form a threshold for the occurrence of some of these ecotypes. During the spring bloom in the Gulf of Aqaba Synechococcus abundances often exceed 105 cells mL−1 (Lindell and Post, 1995; Penno et al., 2006) and members of clade II contribute the bulk of the population (Fuller et al., 2003; Penno et al., 2006), indicating they outcompete the ecotypes that are prevalent during either winter mixing or summer stratification. Of these ecotypes clade I is observed when nutrient concentrations are highest, clades V, VI, and X during transition periods with intermediate nutrient levels and clade III becomes more prevalent during periods of nutrient depletion. These detailed observations over a multiannual period are consistent with earlier studies that spanned a single seasonal cycle (Fuller et al., 2003; Lindell et al., 2005; Penno et al., 2006). Little can be said about the other presumed ecotypes since there are no cultured isolates available and the nutrient physiology of clade X strain RCC307 has been little studied. Clades XI, XIII, and XIV were observed in deeply mixed water masses with higher nutrient concentrations than those observed during times when clade I was present. Clade XII was found in relatively high numbers throughout the seasons. Although more study is required here, its abundance pattern mirrored that of clade II. Clade XII genotypes are closely related to clades II and III (Figure 1) and they seem to form an intermediate ecotype between the two.
Ecotype characterizations have been made from genome comparisons (Palenik et al., 2003; Dufresne et al., 2008; Scanlan et al., 2009). For example, clade III Synechococcus are specialists that occupy oligotrophic niches enabled by their pigment complement (Palenik, 2001), their phosphate scavenging potential and the broad array of N sources they can assimilate (Scanlan et al., 2009). In contrast, the true chromatic adapter CC9311 (clade I) lacks P regulatory and P adaptive genes and contains extra gene copies for ammonium assimilation (Scanlan et al., 2009), consistent with the fluctuating light regimes and nutrient sufficiency in its niche. Gene studies have established a coexistence of nitrate utilizing Synechococcus clades – including novel ecotypes – in different marine waters (Ahlgren and Rocap, 2006; Jenkins et al., 2006). However, Synechococcus WH7803 grows equally fast on nitrate and ammonium (Post, 2005), but strain WH8102 has slower growth rates when utilizing nitrate (Moore et al., 2002). The closely related strains WH7803 (clade IV) and WH7805 (clade V) differ in their urea utilization (Collier et al., 1999). However, strains WH8102 and MIT S9220 with the full set of urea assimilatory genes have different growth efficiencies on this substrate (Moore et al., 2002). It thus seems that ecotypes cannot simply be defined by the presence/absence of genes, but that other factors play into defining the range of ecotype success. In addition to cell external factors like mortality due to phage infection and grazing pressure, cellular processes like transcriptional regulation, turnover of transport and enzyme proteins etc., likely play an important role.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge the opportunity provided by the National Monitoring Program to join monthly cruises in the Gulf of Aqaba and the use of physico-chemical data obtained. This study was enabled through financial support by the EU project MARGENES #QLRT-2001-01226 (Anton F. Post), ISF grant #131/05 (Anton F. Post), and a NATO Science for Peace grant #SfP 982161 (Anton F. Post, Adina Paytan). Pyrosequencing of V6-tags was supported by the International Census of Marine Microorganisms at the MBL, Woods Hole (ICoMM, PI M. L. Sogin). Pyrosequencing data analyses received additional support of the Sloan Foundation. Nina Kamennaya and Mark Chernichovsky provided technical support during sampling and analyses.
References
Ahlgren, N. A., and Rocap, G. (2006). Culture isolation and culture-independent clone libraries reveal new marine Synechococcus ecotypes with distinctive light and N physiologies. Appl. Environ. Microbiol. 72, 7193–7204.
Collier, J., Brahamsha, B., and Palenik, B. (1999). The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amido hydrolase, E.C. 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145, 447–459.
Dufresne, A., Ostrowski, M., Scanlan, D. J., Garczarek, L., Mazard, S., Palenik, B. P., Paulsen, I. T., Tandeau de Marsac, N., Wincker, P., Dossat, C., Ferriera, S., Johnson, J., Post, A. F., Hess, W. R., and Partensky, F. (2008). Unravelling the genomic mosaic of a ubiquitous genus of marine cyanobacteria. Genome Biol. 9, R90.
Durand, M. D. R., Olson, R. J., and Chisholm, S. W. (2001). Phytoplankton population dynamics at the Bermuda Atlantic time series station in the Sargasso Sea. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48, 1983–2003.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Ferris, M. J., and Palenik, B. (1998). Niche adaptation in ocean cyanobacteria. Science 396, 226–228.
Fuller, N. J., Campbell, C., Allen, D. J., Pitt, F. D., Zwirglmaier, K., Le Gall, F., Vaulot, D., and Scanlan, D. J. (2006a). Analysis of photosynthetic picoeukaryote diversity at open ocean sites in the Arabian Sea using a PCR biased towards marine algal plastids. Aquat. Microb. Ecol. 43, 79–93.
Fuller, N. J., Tarran, G. A., Yallop, M., Orcutt, K. M., and Scanlan, D. J. (2006b). Molecular analysis of picocyanobacterial community structure along an Arabian Sea transect reveals distinct spatial separation of lineages. Limnol. Oceanogr. 51, 2515–2526.
Fuller, N. J., Marie, D., Partensky, F., Vaulot, D., Post, A. F., and Scanlan, D. J. (2003). Clade-specific 16S ribosomal DNA oligonucleotides reveal the predominance of a single marine Synechococcus clade throughout a stratified water column in the Red Sea. Appl. Environ. Microbiol. 69, 2430–2443.
Fuller, N. J., Marie, D., Yallop, M., Rivlin, T., West, N. J., Post, A. F., and Scanlan, D. J. (2005). Dynamics of community structure and P status of picocyanobacterial populations in the Gulf of Aqaba, Red Sea during 1999-2000. Limnol. Oceanogr. 50, 363–375.
Garczarek, L., Dufresne, A., Rousvoal, S., West, N., Mazard, S., Marie, D., Claustre, H., Raimbault, P., Post, A. F., Scanlan, D., and Partensky, F. (2007). High vertical and low horizontal microdiversity of Prochlorococcus genotypes in the Mediterranean Sea in summer. FEMS Microbiol. Ecol. 60, 189–206.
Gelman, A., and Rubin, D. B. (1992). Inference from iterative simulation using multiple sequences. Stat. Sci. 7, 434–455.
Genin, A., Lazar, B., and Brenner, S. (1995). Vertical mixing and coral death in the Red Sea following the eruption of mount Pinatubo. Nature 377, 507–510.
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Holmes, R. M., Aminot, A., Kerouel, R., Hooker, B. A., and Peterson, B. J. (1999). A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can. J. Fish. Aquat. Sci. 56, 1801–1808.
Huber, J. A., Mark Welch, D. B., Morrison, H. G., Huse, S. M., Neal, P. R., Butterfield, D. A., and Sogin, M. L. (2007). Microbial population structures in the deep marine biosphere. Science 318, 97–100.
Huelsenbeck, J. P., and Ronquist, F. (2001). MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755.
Huse, S. M., Dethlefsen, L., Huber, J. A., Mark Welch, D. B., Relman, D. A., and Sogin, M. L. (2008). Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 4, e1000255.
Huse, S. M., Huber, J. A., Morrison, H. G., Sogin, M. L., and Mark Welch, D. (2007). Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8, R143.
Jenkins, B. D., Zehr, J. P., Gibson, A., and Campbell, L. (2006). Cyanobacterial assimilatory nitrate reductase gene diversity in coastal and oligotrophic marine environments. Environ. Microbiol. 8, 2083–2095.
Johnson, Z. I., Zinser, E. R., Coe, A., McNulty, N. P., Woodward, E. M. S., and Chisholm, S. W. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311, 1737–1740.
Kamennaya, N. A., Chernichovsky, M., and Post, A. F. (2008). The cyanate utilization capacity of marine unicellular cyanobacteria. Limnol. Oceanogr. 53, 2485–2494.
Kamennaya, N. A., and Post, A. F. (in preparation). Expression of the cyanate acquisition potential among cyanobacterial populations in marine waters.
Kettler, G. C., Martiny, A. C., Huang, K., Zucker, J., Coleman, M. L., Chen, R. S. F., Lapidus, A., Ferriera, S., Johnson, J., Steglich, C., Church, G. M., Richardson, P., and Chisholm, S. W. (2007). Patterns and implications of gene gain and loss in the evolution of Prochlorococcus. PLoS Genet. 3, e231.
Lindell, D., Padan, E., and Post, A. F. (1998). Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180, 1878–1886.
Lindell, D., Penno, S., Al-Qutob, M., David, E., Korpal, T., Lazar, B., and Post, A. F. (2005). Expression of the N-stress response gene ntcA reveals N-sufficient Synechococcus populations in the oligotrophic northern Red Sea. Limnol. Oceanogr. 50, 1932–1944.
Lindell, D., and Post, A. F. (1995). Ultraphytoplankton succession is triggered by deep winter mixing in the Gulf of Aqaba (Eilat), Red Sea. Limnol. Oceanogr. 40, 1130–1141.
Lindell, D., and Post, A. F. (2001). Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl. Environ. Microbiol. 67, 3340–3349.
Mackey, K. R. M., Bristow, L., Parks, D. R., Altabet, M. A., Post, A. F., and Paytan, A. (Submitted). Nitrogen cycling in oligotrophic waters: the influence of light and substrate availability. Prog. Oceanogr.
Marie, D., Brussaard, C. P. D., Thyrhaug, R., Bratbak, G., and Vaulot, D. (1999). Enumeration of marine viruses in culture and natural samples by flow cytometry. Appl. Environ. Microbiol. 65, 45–52.
Martiny, A. C., Tai, A. P., Veneziano, D., Primeau, F., and Chisholm, S. W.. (2008). Taxonomic resolution, ecotypes and the biogeography of Prochlorococcus. Environ. Microbiol. 11, 823–832.
Moore, L. R., Goericke, R., and Chisholm, S. W. (1995). Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence and absorptive properties. Mar. Ecol. Prog. Ser. 116, 259–275.
Moore, L. R., Post, A. F., Rocap, G., and Chisholm, S. W. (2002). Differential nitrogen utilization of the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47, 989–996.
Moore, L. R., Rocap, G., and Chisholm, S. W. (1998). Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393, 464–467.
Olson, R. J., Chisholm, S. W., Zettler, E. R., Altabet, M. A., and Dusenberry, J. A. (1990). Spatial and temporal distribution of prochlorophyte picoplankton in the North Atlantic Ocean. Deep Sea Res. 37, 1033–1051.
Palenik, B. (2001). Chromatic adaptation in marine Synechococcus strains. Appl. Environ. Microbiol. 67, 991–994.
Palenik, B., Brahamsha, B., Larimer, F. W., Land, M., Hauser, L., Chain, P., Lamerdin, J., Regala, W., Allen, E. E., McCarren, J., Paulsen, I., Dufresne, A., Partensky, F., Webb, E. A., and Waterbury, J. (2003). The genome of a motile marine Synechococcus. Nature 424 1037–1042.
Partensky, F., Hess, W. R., and Vaulot, D. (1999). Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63, 106–127.
Penno, S., Lindell, D., and Post, A. F. (2006). Diversity of Synechococcus and Prochlorococcus populations determined from DNA sequences of the N-regulatory gene ntcA. Environ. Microbiol. 8, 1200–1211.
Post, A. F. (2005). “Nutrient limitation of marine cyanobacteria: molecular ecology in an oligotrophic sea,” in Harmful Cyanobacteria, eds J. Huisman, H. C. P. Matthijs, and P. M. Visser (Dordrecht: Kluwer Academic Publishers), 87–107.
Post, A. F., Dedej, Z., Gottlieb, R., Li, H., Thomas, D., El-Absawy, M., El-Naggar, A., El-Gharabawi, M., and Sommer, U. (2002). Spatial and temporal distribution of Trichodesmium spp. in the stratified waters of the Gulf of Aqaba (northern Red Sea). Mar. Ecol. Prog. Ser. 239, 241–250.
Rocap, G., Distel, D. L., Waterbury, J. B., and Chisholm, S. W. (2002). Resolution of Prochlorococcus and Synechococcus ecotypes by using 16S-23S ribosomal DNA internal transcribed space sequences. Appl. Environ. Microbiol. 68, 1180–1191.
Scanlan, D. J., Ostrowski, M., Mazard, S., Dufresne, A., Garczarek, L., Hess, W. R., Post, A. F., Hagemann, M., Paulsen, I., and Partensky, F. (2009). Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73, 249–299.
Sogin, M. L., Morrison, H. G., Huber, J. A., Mark Welch, D., Huse, S. M., Neal, P. R., Arrieta, J. M., and Herndl, G. J. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
Steglich, C., Post, A. F., and Hess, W. R. (2003). Analysis of natural populations of Prochlorococcus spp. in the northern Red Sea using phycoerythrin gene sequences. Environ. Microbiol. 5, 681–690.
Tai, V., and Palenik, B. (2009). Temporal variation of Synechococcus clades at a coastal Pacific Ocean monitoring site. ISME J. 3, 903–915.
Toledo, G., and Palenik, B. (1997). Synechococcus diversity in the California Current as seen by RNA polymerase (rpoC1) gene sequences of isolated strains. Appl. Environ. Microbiol. 63, 4298–4303.
Veldhuis, M. J. W., and Kraay, G. W. (1990). Vertical distribution and pigment composition of a picoplanktonic prochlorophyte in the subtropical North Atlantic: a combined study of HPLC-analysis of pigments and flow cytometry. Mar. Ecol. Prog. Ser. 68, 121–127.
West, N., and Scanlan, D. J. (1999). Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65, 2585–2591.
Wolf-Vecht, A., Paldor, N., and Brenner, S. (1992). Hydrographic indications of advection/convection in the Gulf of Eilat. Deep Sea Res. 39, 1393–1401.
Zinser, E. R., Coe, A., Johnson, Z. I., Martiny, A. C., Fuller, N. J., Scanlan, D. J., and Chisholm, S. W. (2006). Prochlorococcus ecotype abundances in the North Atlantic Ocean as revealed by an improved quantitative PCR method. Appl. Environ. Microbiol. 72, 723–732.
Zwirglmaier, K., Heywood, J. L., Chamberlain, K., Woodward, E. M. S., Zubkov, M. V., and Scanlan, D. J. (2007). Basin-scale distribution patterns of picocyanobacterial lineages in the Atlantic Ocean. Environ. Microbiol. 9, 1278–1290.
Zwirglmaier, K., Jardiller, L., Ostrowski, M., Mazard, S., Garczarek, L., Vaulot, D., Not, F., Massana, R., Ulloa, O., and Scanlan, D. J. (2008). Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ. Microbiol. 10, 147–161.
Keywords: marine cyanobacteria, Synechococcus, succession, diversity, ecotype
Citation: Post AF, Penno S, Zandbank K, Paytan A, Huse SM and Welch DM (2011) Long term seasonal dynamics of Synechococcus population structure in the Gulf of Aqaba, Northern Red Sea. Front. Microbio. 2:131. doi: 10.3389/fmicb.2011.00131
Received: 24 February 2011;
Accepted: 27 May 2011;
Published online: 20 June 2011.
Edited by:
Rex Malmstrom, DOE Joint Genome Institute, USAReviewed by:
Erik Zinser, University of Tennessee, USAJacob Waldbauer, California Institute of Technology, USA
Copyright: © 2011 Post, Penno, Zandbank, Paytan, Huse and Welch. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Anton F. Post, Marine Biological Laboratory, The Josephine Bay Paul Center for Comparative Molecular Biology and Evolution, Woods Hole, MA, USA. e-mail: apost@mbl.edu
