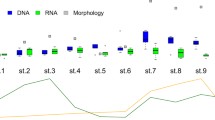
Abstract
Despite the increasing recognition of the quantitative importance of Archaea in all marine systems, the protocols for a rapid estimate of Archaeal diversity patterns in deep-sea sediments have been only poorly tested yet. Sediment samples from 11 deep-sea sites (from 79°N to 36°N, at depths comprised from 469 to 5,571 m) were used to compare the performance of two different primer sets (ARCH21f/ARCH958r and ARCH109f/ARCH 915r) and three restriction enzymes (AluI, Rsa I, and HaeIII) for the fingerprinting analysis of Archaeal diversity using terminal-restriction fragment length polymorphisms (T-RFLP). In silico and experimental analyses indicated that different combinations of primer sets and restriction enzymes provided different values of benthic Archaeal ribotype richness and different Archaeal assemblage compositions. The use of the ARCH109f/ARCH 915r primer set in combination with AluI provided the best results (a number of ribotypes up to four folds higher than other combinations), suggesting that this primer set should be used in future studies dealing with the analysis of the patterns of Archaeal diversity in deep-sea sediments. Multivariate multiple regression analysis revealed that, whatever the T-RFLP protocol utilized, latitude and temperature explained most of the variance in benthic Archaeal ribotype richness, while water depth had a negligible role.

Similar content being viewed by others
References
Baker GC, Smith JJ, Cowan DA (2003) Review and re-analysis of domain-specific 16S primers. J Microbiol Methods 55:541–555
Bent SJ, Forney LJ (2008) The tragedy of the uncommon: understanding limitations in the analysis of microbial diversity. ISME J 2:689–695
Chaban B, Ng SY, Jarrell KF (2006) Archaeal habitats-from the extreme to the ordinary. Can J Microbiol 52:73–116
Chan CO, Claus P, Casper P, Ulrich A, Lueders T, Conrad R (2005) Vertical distribution of structure and function of the methanogenic Archaeal community in Lake Dagow sediment. Environ Microbiol 7:1139–1149
Conrad R, Klose M (2006) Dynamics of the methanogenic Archaeal community in anoxic rice soil upon addition of straw. Eur J Soil Sci 57:476–478
Danovaro R, Luna GM, Dell’Anno A, Pietrangeli B (2006) Comparison of two fingerprinting techniques, terminal restriction fragment length polymorphism and automated ribosomal intergenic spacer analysis, for determination of bacterial diversity in aquatic environments. Appl Environ Microb 72:5982–5989
Danovaro R, Dell’Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C, Weinbauer M (2008) Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454:1084–1087
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci 89:5685–5689
Galand PE, Lovejoy CWF (2006) Remarkably diverse and contrasting Archaeal communities in a large arctic river and the coastal Arctic Ocean. Aquat Microb Ecol 44:115–126
Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J (2005) Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol 71:2303–2309
Hewson I, Steele JA, Capone DG, Fuhrman JA (2006) Remarkable heterogeneity in meso- and bathypelagic bacterioplankton assemblage structure. Limnol Oceanogr 51:1274–1283
Huber JA, Butterfield DA, Baross JA (2002) Temporal changes in Archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl Environ Microbiol 68:1585–1594
Jones SE, Shade AL, McMahon KD, Kent AD (2007) Comparison of primer sets for use in automated ribosomal intergenic spacer analysis of aquatic bacterial communities: an ecological perspective. Appl Environ Microbiol 73:659–662
Kim BS, Oh HM, Kang H, Chun J (2005) Archaeal diversity in tidal flat sediment as revealed by 16S rDNA analysis. J Microbiol 43:144–151
Lanoil BD, La Duc MT, Wright M, Kastner M, Nealson KH, Bartlett D (2005) Archaeal diversity in ODP legacy borehole 892b and associated seawater and sediments of the Cascadia Margin. FEMS Microbiol Ecol 54:167–177
Leuko S, Goh F, Allen MA, Burns BP, Walter MR, Neilan BA (2006) Analysis of intergenic spacer region length polymorphisms to investigate the halophilic archaeal diversity of stromatolites and microbial mats. Extremophiles 11:203–210
Luna GM, Dell’Anno A, Danovaro R (2006) DNA extraction procedure: a critical issue for bacterial diversity assessment in marine sediments. Environ Microbiol 8:308–320
McArdle BH, Anderson MJ (2001) Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82:290–297
Moyer CL, Tiedje JM, Dobbs FC, Karl DM (1996) A computer-simulated restriction fragment length polymorphism analysis of bacterial small-subunit rRNA genes: efficacy of selected tetrameric restriction enzymes for studies of microbial diversity in nature. Appl Environ Microbiol 62:2501–2507
Pernthaler A, Preston CM, Pernthaler J, DeLong EF, Amann R (2002) A comparison of fluorescently labeled oligonucleotide and polynucleotide probes for the detection of pelagic marine bacteria and archaea. Appl Environ Microbiol 68:661–667
Queric NV, Arrieta JS, Soltwedel T, Arntz WE (2008) Prokaryotic community dynamics in the sedimentary microenvironment of the demosponge Tentorium semisuberites from deep Arctic waters. Mar Ecol Prog Ser 370:87–95
Rees GN, Baldwin DS, Watson GO, Perryman S, Nielsen DL (2004) Ordination and significance testing of microbial community composition derived from terminal restriction fragment length polymorphisms: application of multivariate statistics. Anton Leeuw Int J G 86:339–347
Schwarz JI, Eckert W, Conrad R (2006) Community structure of Archaea and Bacteria in a profundal lake sediment in Lake Kinneret (Israel). Syst Appl Microbiol 30:239–254
Shyu C, Soule T, Bent SJ, Foster JA, Forney LJ (2007) MiCA: a web-based tool for the analysis of microbial communities based on terminal-restriction fragment length polymorphisms (T-RFLP) of 16S and 18S rRNA genes. Microb Ecol 53:562–570
Teske A, Sorensen KB (2008) Uncultured Archaea in deep marine subsurface sediments: have we caught them all? ISME J 2:3–18
Vetriani C, Jannasch HW, MacGregor BJ, Stahl DA, Reysenbach A-L (1999) Population structure and phylogenetic characterization of marine benthic Archaea in deep-sea sediments. Appl Environ Microbiol 65:4375–4384
Vetriani C, Tran HV, Kerkhof LJ (2003) Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl Environ Microbiol 69:6481–6488
Winter C, Smit A, Herndl GJ, Weinbauer MG (2004) Impact of virioplankton on Archaeal and Bacterial community richness as assessed in seawater batch cultures. Appl Environ Microbiol 70:804–813
Acknowledgments
This work has been carried out within the frame of the HERMES (Hotspot Ecosystem Research on the Margins of European Seas), EC contract number GOCE-CT-2005-511234, funded by the European Commission’s Framework Six Programme and HERMIONE (Hotspot Ecosystem Research and Man’s Impact on European Seas) funded by the European Commission’s Framework Seven Programme.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luna, G.M., Stumm, K., Pusceddu, A. et al. Archaeal Diversity in Deep-Sea Sediments Estimated by Means of Different Terminal-Restriction Fragment Length Polymorphisms (T-RFLP) Protocols. Curr Microbiol 59, 356–361 (2009). https://doi.org/10.1007/s00284-009-9445-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-009-9445-4