Abstract
During mitosis, all chromosomes must attach to microtubules of the mitotic spindle to ensure correct chromosome segregation. Microtubule attachment occurs at specialized structures at the centromeric region of chromosomes, called kinetochores. These kinetochores can generate microtubule attachments through capture of centrosome-derived microtubules, but in addition, they can generate microtubules themselves, which are subsequently integrated with centrosome-derived microtubules to form the mitotic spindle. Here, we have performed a large scale RNAi screen and identify cyclin G-associated kinase (GAK) as a novel regulator of microtubule generation at kinetochores/chromatin. This function of GAK requires its C-terminal J-domain, which is essential for clathrin recycling from endocytic vesicles. Consistently, cells lacking GAK show strongly reduced levels of clathrin on the mitotic spindle, and reduction of clathrin levels also inhibits microtubule generation at kinetochores/chromosomes. Finally, we present evidence that association of clathrin with the spindle is promoted by a signal coming from the chromosomes. These results identify a role for GAK and clathrin in microtubule outgrowth from kinetochores/chromosomes and suggest that GAK acts through clathrin to control microtubule outgrowth around chromosomes.
Similar content being viewed by others
Introduction
As cells enter mitosis, centrosomes mature through the recruitment of large amounts of γ-tubulin, the major microtubule-nucleating factor, and many other associated proteins (Luders and Stearns 2007). This increase in γ-tubulin at centrosomes likely results in increased microtubule nucleation at centrosomes. In addition to microtubule nucleation at centrosomes, it is now becoming clear that multiple additional sites for microtubule nucleation exist within the mitotic cell. First, several recent studies have shown that microtubule nucleation occurs at pre-existing microtubules in the spindle by the augmin complex, which recruits γ-tubulin to spindle microtubules (Goshima et al. 2008; Lawo et al. 2009; Meireles et al. 2009; Uehara et al. 2009; Wainman et al. 2009; Zhu et al. 2008). Second, chromosomes and kinetochores are also potent sites for microtubule nucleation through the activity of the small GTPase Ran (Clarke and Zhang 2008). Ran is locally activated around chromosomes by its activating protein RCC1 that binds to chromatin. Ran, in turn, activates a plethora of spindle assembly factors (SAFs) by removing the inhibitory effect of importins on these SAFs (Clarke and Zhang 2008). In this way, Ran likely promotes microtubule nucleation and stabilization specifically in the vicinity of the chromatin. These chromosome-derived microtubules are then integrated with centrosome-derived microtubules to form the bipolar spindle (Maiato et al. 2004; Walczak and Heald 2008).
Clathrin is a large trimeric protein that has an essential role in endocytosis. Trimers of clathrin assemble in large multimeric structures at the plasma membrane to facilitate the budding of membranes during endocytosis (Ungewickell and Hinrichsen 2007). After membranes have fully budded, clathrin needs to be removed from vesicles to allow for a new round of vesicle budding. This recycling of clathrin is mediated by the chaperone protein Hsc70 together with auxilin in neuronal and auxilin-2/cyclin G-associated kinase (GAK) in non-neuronal cells (Greener et al. 2000; Ungewickell et al. 1995). Recycling of clathrin is essential for endocytosis, as mouse fibroblasts lacking GAK have severely impaired endocytosis (Lee et al. 2008). In addition to its essential role in endocytosis, clathrin also localizes to the spindle (Okamoto et al. 2000; Royle et al. 2005), and knockdown of clathrin results in defects in chromosome alignment, indicating that clathrin plays an important role in some aspect of spindle function. However, currently, the role of clathrin in the spindle has remained elusive.
Here, we set out to identify novel regulators of spindle assembly using a high-throughput siRNA approach and identify GAK as an important protein for correct spindle assembly and chromosome alignment. Our results show that GAK is required for microtubule growth from chromosomes. Importantly, the mitotic function of GAK could be rescued by expression of full-length GAK, but not by a mutant which is unable to recycle clathrin in interphase. Consistent with this, depletion of GAK completely prevents clathrin recruitment to the spindle, and we show that loss of clathrin also results in defective microtubule outgrowth from chromosomes. Taken together, these results suggest that GAK is needed for chromosome-derived microtubule formation through regulation of clathrin function.
Materials and methods
Cell culture, transfection, and drug treatments
U2OS and Hela cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco) with 6% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. Nocodazole (Sigma) was used at 250 ng/ml.
siRNA and plasmid transfections
The siRNA library that was used for the primary screen was purchased from Ambion. The siRNA library consisting of four siRNAs per gene that was used as a final validation was purchased from Dharmacon. These siRNAs were the “on-target plus” siRNAs that are chemically modified to reduce off-target effects (Jackson et al. 2006). siRNA was transfected using reverse transfection with Hiperfect (Qiagen) according to the manufacturer’s guidelines. Cells were re-transfected after 24 h to improve knockdown efficiency in all experiments, except those described in Fig. 4a, as the combination of plasmid transfection and two siRNA transfection was too toxic for cells. siRNA sequences used here are GAK#2 GCGACACGGUUCUGAAGAU and GAK#3 GGACGCGUGUGACAUUCAA. Clathrin and GAPDH were depleted using an OTP SMARTpool (Dharmacom), and the Hec1 siRNA AAGTTCAAAAGCTGGATGATCTT has been described previously (Tanenbaum et al. 2008). DNA transfections were performed using Fugene 6 (Roche) according to the manufacturer’s guidelines. All GAK plasmids were based on rat GAK and have been described previously (Zhang et al. 2005).
Immunofluorescence
Cells were grown on 10-mm glass coverslips and fixed with 3.7% formaldehyde with 1% triton X-100 and post-fixed with cold methanol. α-tubulin antibody (Sigma) was used 1:7,500, anti-green fluorescent protein (GFP; custom-made) was used 1:500, anti-γ-tubulin (Sigma) was used 1:500, HURP (custom-made (Sillje et al. 2006)) was used 1:500, anti-myc (Covance) and p150glued (BD Bioscience) were used 1:500, anti-CLIP-170 antibody (custom-made (Coquelle et al. 2002)) was used 1:1,000, anti-TPX2 and anti-HSET (Santa Cruz) were used 1:500, anti-Cenp-F (abcam) was used 1:400, and anti-Cenp-E (Gift of Don Cleveland) was used 1:500. Primary antibodies were incubated overnight at room temperature, and secondary antibodies (Alexa 488 and 561, Molecular Probes) were incubated for 1 h at room temperature at a dilution of 1:1,000. DAPI was added before mounting using Vectashield (Vectorlabs). Images acquisition and analysis were performed on a Zeiss LSM510 META confocal microscope (Carl Zeiss) as described previously (Tanenbaum et al. 2009). Brightness and contrast were adjusted with Photoshop 6.0 (Adobe). Images are maximum intensity projections of all Z-planes.
Western blotting
Hela cells were transfected with different siRNAs targeting GAK or with an siRNA targeting GAPDH as a control. Twenty-four hours after transfection cells were re-transfected and 48 h after the first transfection cells were lysed, total GAK levels were analyzed by Western blot using an anti-GAK antibody (MBL international) at 1:1,000 dilutions. Total actin levels were determined using an anti-actin antibody (Tebu bio, dilution 1:2,500) as a loading control.
Results
To identify novel kinases important for spindle assembly, we used a high-throughput RNAi screening approach. In this setup, each human kinase was individually depleted with a pool of three siRNAs. To accomplish this, we made use of a library of 2,148 siRNAs targeting 716 genes, comprising all human kinases. siRNA-mediated depletion was performed in asynchronously growing Hela cells, and 60 h after siRNA transfection, the mitotic index was determined using visual inspection. Wells that had a mitotic index of more than ∼10% were scored as a hit. As expected, depletion of Plk1 resulted in a very high mitotic index (data not shown). In addition, more than 10% of all of the other pools of siRNAs in our library induced a mitotic index of greater than 10%. This high percentage made us suspect that we were picking up a large proportion of off-targets, and therefore, we performed a secondary screen in which the three individual siRNAs targeting each gene that scored positive in the primary screen were tested separately. In this secondary screen, we strictly focused on genes that had not previously been implicated in mitosis. We identified 35 genes for which at least two out of three siRNAs produced a mitotic arrest. Due to the unexpected high rate of suspected off-target hits, we wanted to obtain more stringent validation of our hits and therefore performed a tertiary screen using four additional independent siRNAs that were chemically modified to reduce the chance of off-target effects. From this tertiary screen, we identified only a single kinase, GAK, for which at least three out of four siRNAs resulted in an increased mitotic index (data not shown). This indicates that off-target effects were very frequent in our primary and secondary screen. However, since we were able to reproduce a mitotic arrest with six out of seven oligos targeting GAK, we decided to study the role of GAK in spindle assembly in more detail.
To confirm that GAK was required for normal mitotic progression, we depleted GAK using the six different siRNAs that resulted in a mitotic arrest. Cells depleted of GAK were then fixed, and chromosome alignment was analyzed. Strikingly, all six siRNAs resulted in a substantial increase in the fraction of cells that showed misaligned chromosomes (Fig. 1a), confirming that GAK is required for normal mitotic progression. SiRNAs #2 and #3 reproducibly gave the strongest effects on chromosome alignment; therefore, these siRNAs were used in further experimentation. Importantly, both siRNAs potently reduced GAK protein levels (Fig. 1b). To quantify the severity of the chromosome alignment defects in GAK-depleted cells, the number of misaligned chromosomes per cells was scored. While control cells showed only 0.3 ± 0.1 misaligned chromosomes per cell, this was increased to 3.3 ± 0.7 in cells transfected with GAK siRNA #3 (Fig. 1c). Thus, GAK is not required for the majority of chromosomes to align to the metaphase plate, but in the absence of GAK, chromosome alignment is less efficient. To further confirm a role for GAK in mitosis, we also depleted GAK in U2OS cells. Depletion of GAK in U2OS cells resulted in an even more prominent increase in the percentage of cells with misaligned chromosomes, from 17 ± 6% to 73 ± 15% (Fig. 1d), indicating that the role of GAK in chromosome alignment is not cell type specific. The inability of chromosomes to efficiently align to the metaphase plate could be a consequence of a lack of kinetochore–microtubule attachment, or alternatively, could be due to incorrect (i.e., syntelic) attachments (Kelly and Funabiki 2009). To test whether the misaligned chromosomes in GAK-depleted cells were attached to microtubules, the localization of CLIP-170 and p150glued at kinetochores was analyzed, two markers for unattached kinetochores (Raaijmakers et al. 2009; Tanenbaum et al. 2006). Strikingly, almost all cells with misaligned chromosomes (>80%) showed strong CLIP-170 and p150glued staining at kinetochores of misaligned chromosomes, but not at kinetochores of aligned chromosomes (Fig. 1e). This indicates misaligned chromosomes in GAK-depleted cells have either failed to attach to microtubules or are unable to maintain microtubule attachments, but alignment defects are unlikely due to a failure to release incorrectly attached microtubules. These data suggest that GAK is required for efficient chromosome alignment by facilitating initial kinetochore–microtubule attachment or maintenance of established attachments.
GAK is required for efficient chromosome alignment. a Hela cells were transfected with six different siRNAs targeting GAK or with an siRNA targeting GAPDH as a control. Twenty-four hours after transfection cells were re-transfected and 48 h after the first transfection cells were fixed, DNA was stained with DAPI, and the percentage of mitotic cells with misaligned chromosomes was scored in each condition. b Hela cells were transfected as in (a) with indicated siRNAs, and 48 h after the first transfection cells were lysed and GAK protein levels were determined by western blot. c, d Hela (c) or U2OS cells (d) were treated as in (a), and the average number of misaligned chromosomes was determined (c) or the percentage of mitotic cells with misaligned chromosomes was scored (d). e Hela cells were treated as in (a), but 48 h after transfection cells were fixed and stained for CLIP-170 and p150glued to visualize unattached kinetochores and DAPI was used to stain the DNA. All graphs are averages of three independent experiments with 50 cells scored per condition per experiment. Error bars represent standard deviations. Scale bar in (e) indicates 5 µm
Attachment defects could be a consequence of a direct role for GAK in microtubule capture or stabilization at kinetochores, similar to what has been proposed for kinetochore-associated proteins like the Ska complex, CLIP-170, Cenp-E, and the KMN network (Cheeseman and Desai 2008). Alternatively, defects in kinetochore–microtubule attachment could be a secondary effect of defects in spindle microtubule organization or dynamics. To test this, spindles were analyzed in GAK-depleted cells. In control cells, robust spindles were observed, in which most microtubules grew from spindle poles towards the chromosomes (Fig. 2a). In contrast, in GAK-depleted cells, microtubule staining in the spindle was substantially reduced, especially in the vicinity of chromosomes (Fig. 2a). Furthermore, the pole-to-pole distance of the spindles was also reduced by approximately 25% in the absence of GAK (Fig. 2b). The reduction in pole-to-pole distance was not a secondary effect of the defect in kinetochore–microtubule attachment, as loss of Hec1, which prevents stable kinetochore–microtubule attachments (Martin-Lluesma et al. 2002), did not reduce pole-to-pole distance (Fig. 2b). Consistent with this, the defects in kinetochore–microtubule attachment observed after knockdown of GAK were likely not a consequence of gross defects in kinetochore assembly, as the outer kinetochore components p150glued, Cenp-E, and Cenp-F all accumulated normally at kinetochores in the absence of GAK (Supplemental Fig. S1) Together, these results suggest that GAK has a primary role in bipolar spindle assembly and that defects in spindle assembly in the absence of GAK result in inefficient chromosome alignment.
GAK is required for correct spindle assembly. a U2OS cells were transfected with indicated siRNAs. Twenty-four hours after transfection cells were re-transfected and 48 h after the first transfection cells were fixed so that spindles and DNA could be visualized by anti-α-tubulin staining and DAPI, respectively. b Cells were treated as in (a), and the spindle length was measured using the α-tubulin staining. Graph in (b) shows averages of three independent experiments with 25 cells scored per condition per experiment. Error bars represent standard deviations. Scale bar in (a) indicates 5 µm
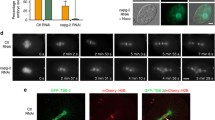
Microtubules are selectively stabilized in the vicinity of chromosomes due to action of the small GTPase Ran, which locally activates microtubule stabilizing and nucleating factors around the chromosomes (Clarke and Zhang 2008). The activity of Ran results in biased growth of microtubules towards the DNA, which in turn leads to a relatively high amount of tubulin polymer near chromosomes. The observation that GAK-depleted cells show reduced microtubule polymer near chromosomes suggest that selective microtubule stabilization in the vicinity of chromosomes is impaired. To test this directly, nocodazole washout experiments were performed (Tulu et al. 2006), in which all microtubules were first depolymerized by the addition of nocodazole, and after removal of nocodazole, microtubule re-growth at both centrosomes and chromosomes was analyzed. In control cells, robust microtubule growth could be observed both at centrosomes and around chromosomes/kinetochores within 3 min after nocodazole removal (Fig. 3a–c). In contrast, in GAK-depleted cells, microtubule outgrowth around chromosomes/kinetochores after nocodazole washout was strongly diminished (Fig. 3a–c). Even 10 min after removal of nocodazole, only very minor amounts of tubulin were observed in the vicinity of chromosomes (Fig. 3a, bottom panels), and spindle assembly was severely delayed. However, microtubule growth from centrosomes appeared normal (Fig. 3a, b), demonstrating that GAK is specifically required for microtubule outgrowth around chromosomes. These results explain why GAK-depleted spindles have reduced microtubule density in the vicinity of chromosomes and suggest that GAK is important for Ran-dependent microtubule nucleation/stabilization.
GAK is required for the generation of chromatin-derived microtubules. a–c U2OS cells were transfected with indicated siRNAs. Twenty-four hours after transfection, cells were re-transfected, and 48 h after the first transfection, cells were treated with nocodazole for 6 h, washed four times with medium to remove nocodazole, and fixed after 3 min (a, upper panels and b, c) or after 10 min (a, lower panels) and stained for α-tubulin and γ-tubulin. DAPI was used to visualize the DNA. a Representative images. b, c Quantifications. d Cells were transfected as in (a–c) but were fixed after 48 h without nocodazole treatment. Cells were then stained for HURP and α-tubulin, and DNA was stained with DAPI. HURP localizes specifically to DNA-proximal microtubules both in control cells and in GAK-depleted cells. Graphs in (b, c) show averages of three independent experiments with 25 cells scored per condition per experiment. Error bars represent standard deviations. Scale bars in (a, d) indicate 5 µm
To better understand how GAK might control microtubule outgrowth from chromosomes, we attempted to localize GAK during mitosis, but did not observe any specific localization to microtubules or chromosomes (data not shown). This suggests that either the role of GAK in spindle assembly is indirect or that the epitope recognized by our antibody is masked by association of GAK with the spindle. Either way, since we observed that GAK is needed for chromosome-dependent microtubule growth, we wondered if GAK could control the establishment or maintenance of the Ran signaling gradient that is known to play a role in chromosome-dependent microtubule nucleation. To test this, control cells and cells depleted of GAK were stained for the Ran target protein HURP. HURP binds to microtubules specifically in the vicinity of chromosomes, because its microtubule-binding affinity is controlled by the chromatin-derived Ran signal (Koffa et al. 2006; Sillje et al. 2006). Indeed, in control cells, HURP clearly bound to DNA-proximal microtubules (Fig. 3d). However, in GAK-depleted cells HURP still localized to microtubules specifically near chromosomes (Fig. 3d), suggesting that the Ran signaling gradient is still functioning in the absence of GAK. Similarly, the Ran targets HSET and TPX2 also localized normally to the spindle in the absence of GAK (Supplemental Fig. S2). Therefore, GAK is most likely not an upstream regulator of Ran. Taken together, these results show that GAK is essential for microtubule outgrowth from the chromosomes and suggest that it acts downstream of Ran signaling.
In interphase, GAK promotes the uncoating of clathrin from endocytic vesicles through recruitment of the chaperone protein Hsc70 (Greener et al. 2000; Zhang et al. 2005). For this activity, GAK requires its C-terminal J-domain, but its kinase domain is dispensable (Greener et al. 2000; Zhang et al. 2005). Interestingly, like GAK, clathrin is required for efficient chromosome alignment (Royle et al. 2005), suggesting that GAK might regulate chromosome alignment via clathrin. Therefore, we performed a series of RNAi rescue experiments, to test whether the function of GAK during mitosis is related to its function in the uncoating of clathrin-coated vesicles. In control Hela cells, almost 80% of mitotic cells were in metaphase, while the remaining ∼20% of cells were in prometaphase (Fig. 4a). In contrast, when GAK was depleted, many cells showed a tight metaphase plate with a few chromosomes that were distal to the metaphase plate, in the vicinity of spindle poles. This type of configuration was easily distinguishable from normal prometaphase cells and was observed in control cells only at very low frequency (<5% of cells). A strong increase in cells with polar chromosomes was observed in GAK-depleted cells, and this could be rescued by expression of an siRNA-insensitive form of GAK (Fig. 4a). Similarly, a kinase dead version of GAK was able to fully restore chromosome alignment in absence of GAK, indicating that the kinase activity of GAK is not required for proper chromosome alignment. However, when the J-domain was removed from GAK, it could no longer restore normal chromosome alignment in GAK-depleted cells (Fig. 4a), suggesting that the major function of GAK is to recycle clathrin from endocytic vesicles at the onset of mitosis to allow it to function at the spindle.
GAK mediates clathrin recruitment to the spindle. a Hela cells were transfected with indicated myc-tagged plasmids and 24 h later, cells were transfected with GAK siRNA where indicated. Forty-eight hours after siRNA transfection cells were fixed and stained for myc. DAPI was used to visualize the DNA. Chromosome alignment was then analyzed in mitotic cells expressing the indicated plasmids. b, c U2OS cells stably expressing green fluorescent protein (GFP)-clathrin light chain were transfected with indicated siRNAs. Twenty-four hours after transfection cells were re-transfected and 48 h after the first transfection cells were fixed and stained for α-tubulin and GFP. DAPI was used to visualize the DNA. a Representative cells. b Quantifications. d–g U2OS cells were transfected with indicated siRNAs. Twenty-four hours after transfection cells were re-transfected and 60 h after the first transfection cells were treated with nocodazole for 6 h, washed four times to remove nocodazole, fixed after 3 min, and stained for α-tubulin and γ-tubulin. DAPI was used to visualize the DNA. The amount of microtubules around centrosomes (visualized by γ-tubulin staining; d) or chromosomes (visualized by DAPI staining; e) was then analyzed. Images in (f) show a control cell (upper panel) and a clathrin heavy chain-depleted cell with a strong phenotype (lower panel). Graphs in (b–e, g) show averages of three independent experiments with 25 cells scored per condition per experiment in (c–e, g) and 50 cells per condition in (c). Error bars represent standard deviations. Scale bars in (a, f) indicate 5 µm
To determine whether GAK is required for clathrin function during mitosis, we examined clathrin localization in mitotic cells. During mitosis, clathrin was shown to localize to the spindle (Okamoto et al. 2000; Royle et al. 2005). Unfortunately, due to high variability in clathrin staining on the spindle, we were unable to quantify this staining, and therefore, we generated a stable cell line expressing GFP-clathrin light chain (CLC), which robustly localized to spindles (Fig. 4b). Strikingly, GFP-CLC did not fully overlap with α-tubulin staining, but concentrated on microtubules specifically in the vicinity of chromosomes (Fig. 4b), suggesting that, like HURP, clathrin binding to spindle microtubules is regulated by Ran signaling. When cells were depleted of GAK, GFP-CLC was no longer observed along spindle microtubules (Fig. 4b, c), confirming that GAK is indeed essential for clathrin localization during mitosis.
If GAK indeed acts by modulating clathrin function, knockdown of clathrin is also expected to result in a decrease in microtubule outgrowth from chromosomes. To test this, nocodazole washout experiments were performed as described above in either control cells or cells treated with clathrin heavy chain (CHC) siRNA. Indeed, knockdown of clathrin reduced the amount of microtubules growing out from chromosomes, but not from centrosomes in this assay, further suggesting that GAK acts through clathrin to promote spindle assembly (Fig. 4d–f). Finally, to test whether GAK and clathrin act in the same pathway, GAK and CHC were co-depleted from cells, and nocodazole washout experiments were again performed. Cells depleted of both GAK and CHC did not show an increased defect in microtubule outgrowth from chromosomes when compared to depletion of GAK alone (Fig. 4g). These results indicated that GAK and clathrin act in a common pathway and further suggest that GAK controls spindle assembly through clathrin.
Discussion
Here, we identify GAK as a novel factor essential for efficient spindle assembly and chromosome alignment. GAK has a well-studied role in recycling clathrin from clathrin-coated vesicles in interphase, and we now show that GAK is also required for efficient spindle assembly. We show that GAK recruits clathrin to the spindle and that both GAK and clathrin promote outgrowth of microtubules from chromosomes. In addition, we find that the alignment defect of GAK-depleted cells can only be reverted by variants of GAK that are able to recycle clathrin from endosomes. Moreover, we show that GAK and clathrin are both essential for chromosome-dependent microtubule outgrowth, providing a mechanistic explanation for their contribution to chromosome alignment. Taken together, our results suggest that the primary function of GAK is to recycle clathrin from vesicles, thereby enabling clathrin to target to the spindle and promote chromosome-dependent microtubule outgrowth.
These results are consistent with a very recent report, which also showed chromosome alignment defects and inhibition of clathrin recruitment to the spindle after GAK depletion (Shimizu et al. 2009). However, Shimizu et al. found that centrosomes are often fragmented in cells depleted of GAK, a phenotype that was not observed in our study. One possible explanation for this difference was the use of different cell lines. Shimizu et al. used Hela cells in their study, and we have observed that in some clones of Hela cells, centrosomes fragment after prolonged mitotic arrest (unpublished observation), while this is not the case in U2OS cells.
Clathrin is an essential component of the endocytic pathway. It is recruited to membranes during endocytosis and forms basket-like structures around vesicles as they bud from the plasma membrane. Once vesicles have budded from the membrane, clathrin is removed from vesicles by GAK and Hsc70 to allow for it to be re-used (Greener et al. 2000). Here, we show that in GAK-depleted cells, clathrin no longer localizes to the spindle, suggesting that GAK-dependent recycling of clathrin is also required to allow clathrin to function in mitosis. The notion that GAK promotes spindle recruitment of clathrin primarily through its role in clathrin recycling at vesicles in interphase is also supported by the fact that the J-domain of GAK, which is essential for clathrin recycling, is also essential for the mitotic function of GAK. Nonetheless, we cannot completely exclude that besides its role in clathrin recruitment to the spindle, GAK might also have additional functions in spindle assembly. In addition, loss of GAK perturbs endocytosis during interphase. It is possible that cellular dysfunction due to the block in endocytosis somehow contributes to the spindle defects as well. We feel this is unlikely to fully explain the spindle defects in GAK-depleted cells, because the interphase and mitotic of clathrin can, at least in part, be uncoupled (Royle and Lagnado 2006). This indicates that spindle defects observed in the absence of clathrin cannot be completely attributed to defects in endocytosis.
Although it was shown several years ago that clathrin is required for efficient chromosome alignment (Royle et al. 2005), its molecular function within the spindle has remained elusive. Here, we show that loss of clathrin results in reduced microtubule outgrowth from the chromosomes, suggesting that clathrin might be important downstream of Ran to promote microtubule nucleation or stabilization in the vicinity of the chromatin. This idea is supported by the fact that GFP-CLC localizes more prominently to spindle microtubules near the DNA than at the spindle poles. A very similar localization has been described for the Ran target HURP (Sillje et al. 2006; Wong and Fang 2006), suggesting that clathrin might be a Ran target as well. Surprisingly, this specific localization of clathrin has not been reported previously for endogenous clathrin, and we have unfortunately not been able to reliably detect endogenous clathrin in this study. However, it is interesting to note that endogenous clathrin was reported to localize specifically to kinetochore–microtubules, but not to astral microtubules, which are far away from the chromosomes (Royle et al. 2005), supporting the hypothesis that clathrin associated with the spindle is regulated by a signal emanating from the chromosomes. It will therefore be important to test whether clathrin binding to the spindle is indeed dependent on Ran signaling. Future work will hopefully answer this question, perhaps using a system in which Ran-dependent spindle assembly can be studied more easily, like cell-free Xenopus egg extracts.
As there is no evidence that either GAK or clathrin has direct microtubule-binding affinity, it is not evident how these proteins might promote microtubule outgrowth from chromosomes. Since we favor a model in which GAK acts mainly through clathrin, the question becomes, how does clathrin promote microtubule outgrowth? One possibility is that clathrin acts as a scaffold for microtubule stabilizing proteins near the DNA. If clathrin localization were dependent on Ran signaling, this model would predict that Ran could act by generating a clathrin gradient around chromosomes, which in turn would promote microtubule nucleation or stabilization through recruitment of microtubule-binding proteins. Hopefully, future work will identify novel binding partners of clathrin in spindle assembly to further elucidate its role in chromosome-dependent microtubule growth.
References
Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 9:33–46
Clarke PR, Zhang C (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 9:464–477
Coquelle FM, Caspi M, Cordelieres FP, Dompierre JP, Dujardin DL, Koifman C, Martin P, Hoogenraad CC, Akhmanova A, Galjart N et al (2002) LIS1, CLIP-170’s key to the dynein/dynactin pathway. Mol Cell Biol 22:3089–3102
Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD (2008) Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol 181:421–429
Greener T, Zhao X, Nojima H, Eisenberg E, Greene LE (2000) Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J Biol Chem 275:1365–1370
Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K et al (2006) Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. Rna 12:1197–1205
Kelly AE, Funabiki H (2009) Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 21:51–58
Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW (2006) HURP is part of a Ran-dependent complex involved in spindle formation. Curr Biol 16:743–754
Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B et al (2009) HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol 19:816–826
Lee DW, Zhao X, Yim YI, Eisenberg E, Greene LE (2008) Essential role of cyclin-G-associated kinase (Auxilin-2) in developing and mature mice. Mol Biol Cell 19:2766–2776
Luders J, Stearns T (2007) Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol 8:161–167
Maiato H, Rieder CL, Khodjakov A (2004) Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J Cell Biol 167:831–840
Martin-Lluesma S, Stucke VM, Nigg EA (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297:2267–2270
Meireles AM, Fisher KH, Colombie N, Wakefield JG, Ohkura H (2009) Wac: a new Augmin subunit required for chromosome alignment but not for acentrosomal microtubule assembly in female meiosis. J Cell Biol 184:777–784
Okamoto CT, McKinney J, Jeng YY (2000) Clathrin in mitotic spindles. Am J Physiol Cell Physiol 279:C369–C374
Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH (2009) RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci 122:2436–2445
Royle SJ, Lagnado L (2006) Trimerisation is important for the function of clathrin at the mitotic spindle. J Cell Sci 119:4071–4078
Royle SJ, Bright NA, Lagnado L (2005) Clathrin is required for the function of the mitotic spindle. Nature 434:1152–1157
Shimizu H, Nagamori I, Yabuta N, Nojima H (2009) GAK, a regulator of clathrin-mediated membrane traffic, also controls centrosome integrity and chromosome congression. J Cell Sci 122:3145–3152
Sillje HH, Nagel S, Korner R, Nigg EA (2006) HURP is a Ran-importin beta-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16:731–742
Tanenbaum ME, Galjart N, van Vugt MA, Medema RH (2006) CLIP-170 facilitates the formation of kinetochore-microtubule attachments. Embo J 25:45–57
Tanenbaum ME, Macurek L, Galjart N, Medema RH (2008) Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. Embo J 27:3235–3245
Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH (2009) Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol 19:1703–1711
Tulu US, Fagerstrom C, Ferenz NP, Wadsworth P (2006) Molecular requirements for kinetochore-associated microtubule formation in mammalian cells. Curr Biol 16:536–541
Uehara R, Nozawa RS, Tomioka A, Petry S, Vale RD, Obuse C, Goshima G (2009) The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc Natl Acad Sci U S A 106:6998–7003
Ungewickell EJ, Hinrichsen L (2007) Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol 19:417–425
Ungewickell E, Ungewickell H, Holstein SE, Lindner R, Prasad K, Barouch W, Martin B, Greene LE, Eisenberg E (1995) Role of auxilin in uncoating clathrin-coated vesicles. Nature 378:632–635
Wainman A, Buster DW, Duncan T, Metz J, Ma A, Sharp D, Wakefield JG (2009) A new Augmin subunit, Msd1, demonstrates the importance of mitotic spindle-templated microtubule nucleation in the absence of functioning centrosomes. Genes Dev 23:1876–1881
Walczak CE, Heald R (2008) Mechanisms of mitotic spindle assembly and function. Int Rev Cytol 265:111–158
Wong J, Fang G (2006) HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J Cell Biol 173:879–891
Zhang CX, Engqvist-Goldstein AE, Carreno S, Owen DJ, Smythe E, Drubin DG (2005) Multiple roles for cyclin G-associated kinase in clathrin-mediated sorting events. Traffic 6:1103–1113
Zhu H, Coppinger JA, Jang CY, Yates JR 3rd, Fang G (2008) FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J Cell Biol 183:835–848
Acknowledgements
We thank Erich Nigg for the anti-HURP antibody and Don Cleveland for anti-Cenp-E antibody. We also thank the other members of our research groups for helpful discussions and suggestions and Livio Kleij for support with microscopy. We would also like to thank the Biomedicum Helsinki Molecular Imaging Unit for services. This work was supported by Academy of FInland and EU FP6 Program (ENFIN), LSHG-CT-2005-518254. This work was further supported by the Division for Earth and Life Sciences (ALW) and the Netherlands Genomic Initiative (NGI), both with financial aid of the Netherlands Organization for Scientific Research (NWO).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure S1
No defects in kinetochore assembly are observed after GAK depletion. a–c U2OS cells were transfected with either GAPDH or GAK siRNAs. Cells were re-transfected after 24 and 48 h after the first transfection cells were treated with nocodazole for 6 h to depolymerize all microtubules. Cells were then fixed and stained with indicated antibodies, and the DNA was stained with DAPI. Scale bars indicate 5 µm. (GIF 202 kb)
Supplemental Figure S2
TPX2 and HSET localize normally in GAK-depleted cells. a, b U2OS cells were transfected with either GAPDH or GAK siRNAs. Cells were re-transfected after 24 and 48 h after the first transfection cells were fixed and stained with indicated antibodies, and the DNA was stained with DAPI. Scale bars indicate 5 µm. (GIF 164 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tanenbaum, M.E., Vallenius, T., Geers, E.F. et al. Cyclin G-associated kinase promotes microtubule outgrowth from chromosomes during spindle assembly. Chromosoma 119, 415–424 (2010). https://doi.org/10.1007/s00412-010-0267-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-010-0267-8