Abstract
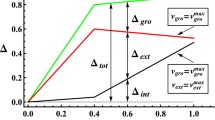
Flux balance analysis, based on the mass conservation law in a cellular organism, has been extensively employed to study the interplay between structures and functions of cellular metabolic networks. Consequently, the phenotypes of the metabolism can be well elucidated. In this paper, we introduce the Expanded Flux Variability Analysis (EFVA) to characterize the intrinsic nature of metabolic reactions, such as flexibility, modularity and essentiality, by exploring the trend of the range, the maximum and the minimum flux of reactions. We took the metabolic network of Escherichia coli as an example and analyzed the variability of reaction fluxes under different growth rate constraints. The average variability of all reactions decreases dramatically when the growth rate increases. Consider the noise effect on the metabolic system, we thus argue that the microorganism may practically grow under a suboptimal state. Besides, under the EFVA framework, the reactions are easily to be grouped into catabolic and anabolic groups. And the anabolic groups can be further assigned to specific biomass constitute. We also discovered the growth rate dependent essentiality of reactions.
Similar content being viewed by others
References
Reed J L, Vo T D, Schilling C H, et al. An expanded genome-scale model of Escherichia coli K-12 (iJR904 (GSM/GPR). Genome Biol, 2003, 4: R54
Duarte N C, Herrgard M J, Palsson B O. Reconstruction and validation of saccharomyces cerevisiae iND750, a fully compart-mentalized genome-scale metabolic model. Genome Res, 2004, 14: 1298–1309
Edwards J S, Ibarra R U, Palsson B O. In silico pre- dictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nature Biotech, 2001, 19: 125–130
Segre D, Vitkup D, Church G M. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci USA, 2002, 99: 15112–15117
Edwards J S, Covert M, Palsson B O. Metabolic modelling of microbes: the flux-balance approach. Environ Microbiol, 2002, 4: 133–140
Price N D, Papin J A, Schilling C H, et al. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotech, 2003, 21: 162–169
Stelling J, Sauer U, Szallasi Z, et al. Robustness of cellular Functions. Cell, 2004, 118: 675–685
Reed J L, Palsson B O. Genome-scale in silico models of E. coli have multiple equivalent phenotypic states: Assessment of coorelated reaction subsets that comprise network states. Genome Res, 2004, 14: 1797–1805
Burgard A P, Nikolaev E V, Schilling C H, et al. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res, 2004, 14: 301–312
Klamt S, Gilles E D. Minimal cut sets in biochemical reaction networks. Bioinformatics, 2004, 20: 226–234
Edwards J S, Palsson B O. The Escherichia coli MG1655 in silico metabolic genotype: Its definition, characteristics, and capabilities. Proc Natl Acad Sci USA, 2000, 297: 5528–5533
Mahadevan R, Schilling C H. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng, 2003, 5: 264–276
Famili I, Foster J, Nielsen J, et al. Saccharomyces cerevisiae phenotypes can be predicted by using constraint-based analysis of a genome-scale reconstructed metabolic network. Proc Natl Acad Sci USA, 2003, 100: 13134–13139
Schuster S, Fell D A, Dandekar T. A general definition of metabolic pathways useful for systematic organization and analysis of complex metabolic networks. Nature Biotech, 2000, 18: 326–332
Edwards J S, Pallson B O. Robustness analysis of the Escherichia coli metabolic network. Biotech Prog, 2000, 16: 927–939
Bilu Y, Shlomi T, Barkai N, et al. Conservation of expression and sequence of metabolic genes is reflected by activity across metabolic states. Plos Comput Biol, 2007, 2: 932–938
Wiback S, Famili I, Greenberg H, et al. Monte Carlo sampling can be used to determine the size and shape of the steady-state flux space. J Theoret Biol, 2004, 228: 437–447
Price N, Reed J, Papin J, et al. Network-based analysis of metabolic regulation in the human red blood cell. J Theoret Biol, 2003, 225: 185–194
Fong S, Joyce A, Palsson B. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res, 2005, 15: 1365–1372
Fischer E, Sauer U. Large-scale in vivo flux analysis shows rigidity and suboptimal performance of Bacillus subtilis metabolism. Nature Genet, 2005, 37: 636–640
Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science, 2002, 292: 504–507
Kitano H. Biological robustness. Nature Rev Genet, 2004, 5: 826–837
Stelling J, Klamt S, Bettenbrock K, et al. Metabolic network structure determines key aspects of fuctionality and regulation. Nature, 2002, 420: 190–193
Meyers L A, Lee J F, Perthwaite M, et al. The Robustness of naturally and artificially selected nucleic acid secondary structures. J Mol Evolut, 2004, 58: 681–691
Novak M, Pfeiffer T, Lenski R E, et al. Experimental tests for an evolutionary trade-off between growth rate and yield in E. coli. Am Nat, 2006, 168: 242–251
Caste M E, Doyle J. Bow ties, metabolism and disease. Trends Biotech, 2004, 22: 446–450
Papin P A, Reed J L, Palsson B O. Hierarchial thinking in network biology: the unbiased modularization of biochemical networks. Trends Biochem Sci, 2004, 29: 641–647
Guimera R, Amaral A N. Functional cartography of complex metabolic networks. Nature, 2005, 433: 895–900
Hartwell L H, Hopfield J J, Leibler S, et al. From molecular to modular cell biology. Nature, 1999, 402: C47–C52
Papin J A, Price N D, Wiback S J, et al. Metabolic pathways in the post-genome era. Trends Biochem Sci, 2003, 28: 250–258
Schuster S, Dandekar T, Fell D A. Detection of elementary flux mode in biochemical networks: A promising tool for pathway analysis and metabolic engineering. Trends Biochem Sci, 1999, 17: 53–60
Schilling C H, Cover M W, Famili I, et al. Genome-scale metabolic model of Helicobacter pylori. J Bacteriol, 2002, 184: 4582–4593
Samal A, Singh S, Giri V, et al. Low degree metabolites explain essential reactions and enhance modularity in biological networks. BMC Bioinformatics, 2006, 7: 118–127
Mahadevan R, Palsson B O. Properties of metabolic networks: Structure vs. function. Biophys J, 2005, 88: L7–L9
Jeong H, Tombor B, Albert R, et al. The large-scale organization of metabolic network. Nature, 2000, 407: 651–654
Jeong H, Mason S P, Barabasi A L, et al. Lethality and centrality in protein networks. Nature, 2001, 411: 41–42
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the National Natural Science Foundation of China (Grant No. 10721403), National Basic Research Program of China (Grant Nos. 2006CB910706, 2007CB814800, 2009CB918500), Chun-Tsung endowment at Peking University and National Fund for Fostering Talents of Basic Science (Grant No. J0630311)
Electronic supplementary material
About this article
Cite this article
Chen, T., Xie, Z. & Ouyang, Q. Expanded flux variability analysis on metabolic network of Escherichia coli . Chin. Sci. Bull. 54, 2610–2619 (2009). https://doi.org/10.1007/s11434-009-0341-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11434-009-0341-x