Abstract
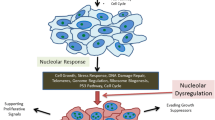
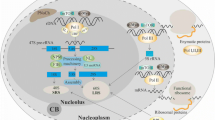
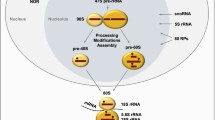
Nucleoli perform a crucial cell function, ribosome biogenesis, and of critical relevance to the subject of this review, they are also extremely sensitive to cellular stresses, which can cause loss of function and/or associated structural disruption. In recent years, we have learned that cells take advantage of this stress sensitivity of nucleoli, using them as stress sensors. One major protein regulated by this role of nucleoli is the tumor suppressor p53, which is activated in response to diverse cellular injuries in order to exert its onco-protective effects. Here we discuss a model of nucleolar regulation of p53, which proposes that key steps in the promotion of p53 degradation by the ubiquitin ligase MDM2 occur in nucleoli, thus providing an explanation for the observed link between nucleolar disruption and p53 stability. We review current evidence for this compartmentalization in p53 homeostasis and highlight current limitations of the model. Interestingly, a number of current chemotherapeutic agents capable of inducing a p53 response are likely to do so by targeting nucleolar functions and these compounds may serve to inform further improved therapeutic targeting of nucleoli.



Similar content being viewed by others
References
von Bertalanffy L (1960) Problems of life: an evaluation of modern biological and scientific thought. Harper, New York
Warner JR, McIntosh KB (2009) How common are extraribosomal functions of ribosomal proteins? Mol Cell 34(1):3–11. doi:10.1016/j.molcel.2009.03.006
Olson MO, Hingorani K, Szebeni A (2002) Conventional and nonconventional roles of the nucleolus. Int Rev Cytol 219:199–266. doi:10.1016/S0074-7696(02)19014-0
Pederson T, Tsai RYL (2009) In search of nonribosomal nucleolar protein function and regulation. J Cell Biol 184(6):771–776. doi:10.1083/jcb.200812014
Vazquez A, Bond EE, Levine AJ, Bond GL (2008) The genetics of the p53 pathway, apoptosis and cancer therapy. Nat Rev Drug Discov 7(12):979–987. doi:10.1038/nrd2656
Zilfou JT, Lowe SW (2009) Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol 1(5):a001883. doi:10.1101/cshperspect.a001883
Meek DW (2009) Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 9(10):714–723. doi:10.1038/nrc2716
Bensaad K, Tsuruta A, Selak MA, Vidal MNC, Nakano K, Bartrons R, Gottlieb E, Vousden KH (2006) TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126(1):107–120. doi:10.1016/j.cell200605036
Appella E, Anderson CW (2001) Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 268:2764–2772. doi:10.1046/j.1432-1327.2001.02225.x
Ljungman M (2000) Dial 9-1-1 for p53: mechanisms of p53 activation by cellular stress. Neoplasia 2:208–225
Lane D, Lain S (2002) Therapeutic exploitation of the p53 pathway. Trends Mol Med 8:S38–S42. doi:10.1016/S1471-4914(02)02309-2
Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP (2009) Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer 9(12):862–873. doi:10.1038/nrc2763
Vlatkovic N, Crawford K, Rubbi CP, Boyd MT (2011) Tissue-specific therapeutic targeting of p53 in cancer: one size does not fit all. Curr Pharm Des 17(6):618–630. doi:10.2174/138161211795222568
Senderowicz AM (2004) Targeting cell cycle and apoptosis for the treatment of human malignancies. Curr Opin Cell Biol 16(6):670–678. doi:10.1016/j.ceb.2004.09.014
Marine JC, Lozano G (2010) Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ 17(1):93–102. doi:10.1038/cdd.2009.68
Freedman DA, Wu L, Levine AJ (1999) Functions of the MDM2 oncoprotein. Cell Mol Life Sci 55(1):96–107. doi:10.1007/s000180050273
Carter S, Vousden KH (2008) p53-Ubl fusions as models of ubiquitination, sumoylation and neddylation of p53. Cell Cycle 7(16):2519–2528. doi:10.4161/cc.7.16.6422
Carter S, Bischof O, Dejean A, Vousden KH (2007) C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol 9:428–435. doi:10.1038/ncb1562
Xirodimas DP (2008) Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans 036(5):802–806. doi:10.1042/BST0360802
Zhang Y, Xiong Y (2001) Control of p53 ubiquitination and nuclear export by MDM2 and ARF. Cell Growth Differ 12(4):175–186
Rubbi CP, Milner J (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J 22:6068–6077. doi:10.1093/emboj/cdg579
Horn HF, Vousden KH (2004) Cancer: guarding the guardian? Nature 427:110–111. doi:10.1038/427110a
Olson MO (2004) Sensing cellular stress: another new function for the nucleolus? Sci STKE 2004(224):pe10. doi:10.1126/stke.2242004pe10
Rudra D, Warner JR (2004) What better measure than ribosome synthesis? Genes Dev 18:2431–2436. doi:10.1101/gad.1256704
Boulon S, Westman BJ, Hutten S, Boisvert F-M, Lamond AI (2010) The nucleolus under stress. Mol Cell 40(2):216–227. doi:10.1016/j.molcel.2010.09.024
Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, Kellner M, Gruber-Eber A, Kremmer E, Hölzel M, Eick D (2010) Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem 285(16):12416–12425. doi:10.1074/jbc.M109.074211
Chan PK (1992) Characterization and cellular localization of nucleophosmin/B23 in HeLa cells treated with selected cytotoxic agents (studies of B23-translocation mechanism). Exp Cell Res 203:174–181. doi:10.1016/0014-4827(92)90053-B
Chan PK, Qi Y, Amley J, Koller CA (1996) Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett 100:191–197. doi:10.1016/0304-3835(95)04100-1
Hadjiolov AA (1985) The nucleolus and ribosome biogenesis. Springer, Wien
Hernandez-Verdun D (2006) Nucleolus: from structure to dynamics. Histochem Cell Biol 125:127–137. doi:10.1007/s00418-005-0046-4
Sirri V, Urcuqui-Inchima S, Roussel P, Hernandez-Verdun D (2008) Nucleolus: the fascinating nuclear body. Histochem Cell Biol 129(1):13–31. doi:10.1007/s00418-007-0359-6
Sirri V, Hernandez-Verdun D, Roussel P (2002) Cyclin-dependent kinases govern formation and maintenance of the nucleolus. J Cell Biol 156:969–981. doi:10.1083/jcb.200201024
Louvet E, Junera HR, Berthuy I, Hernandez-Verdun D (2006) Compartmentation of the nucleolar processing proteins in the granular component is a CK2-driven process. Mol Biol Cell 17:2537–2546. doi:10.1091/mbc.E05-10-0923
Sirri V, Roussel P, Hernandez-Verdun D (2000) In vivo release of mitotic silencing of ribosomal gene transcription does not give rise to precursor ribosomal RNA processing. J Cell Biol 148:259–270. doi:10.1083/jcb.148.2.259
Digman MA, Brown CM, Sengupta P, Wiseman PW, Horwitz AR, Gratton E (2005) Measuring fast dynamics in solutions and cells with a laser scanning microscope. Biophys J 89(2):1317–1327. doi:10.1529/biophysj.105.062836
Mayer C, Grummt I (2005) Cellular stress and nucleolar function. Cell Cycle 4:1036–1038. doi:10.4161/cc.4.8.1925
Grimm T, Hölzel M, Rohrmoser M, Harasim T, Malamoussi A, Gruber-Eber A, Kremmer E, Eick D (2006) Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res 34(10):3030–3043. doi:10.1093/nar/gkl378
Pestov DG, Strezoska Z, Lau LF (2001) Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol 21:4246–4255. doi:10.1128/MCB.21.13.4246-4255.2001
Hölzel M, Orban M, Hochstatter J, Rohrmoser M, Harasim T, Malamoussi A, Kremmer E, Längst G, Eick D (2010) Defects in 18 S or 28 S rRNA processing activate the p53 pathway. J Biol Chem 285(9):6364–6370. doi:10.1074/jbc.M109.054734
Rohrmoser M, Hölzel M, Grimm T, Malamoussi A, Harasim T, Orban M, Pfisterer I, Gruber-Eber A, Kremmer E, Eick D (2007) Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol 27(10):3682–3694. doi:10.1128/mcb.00172-07
Yuan X, Zhou Y, Casanova E, Chai M, Kiss E, Gröne H-J, Schütz G, Grummt I (2005) Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol Cell 19(1):77–87
Zhao J, Yuan X, Frödin M, Grummt I (2003) ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA Polymerase I transcription and cell growth. Mol Cell 11(2):405–413. doi:10.1016/S1097-2765(03)00036-4
White RJ (2005) RNA polymerases I and III, growth control and cancer. Nat Rev Mol Cell Biol 6(1):69–78
Drygin D, Rice WG, Grummt I (2010) The RNA Polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu Rev Pharmacol Toxicol 50(1):131–156. doi:10.1146/annurev.pharmtox.010909.105844
Grewal SS, Li L, Orian A, Eisenman RN, Edgar BA (2005) Myc-dependent regulation of ribosomal RNA synthesis during Drosophila development. Nat Cell Biol 7:295–302
Grandori C, Gomez-Roman N, Felton-Edkins ZA, Ngouenet C, Galloway DA, Eisenman RN, White RJ (2005) c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol 7:311–318
Arabi A, Wu S, Ridderstrale K, Bierhoff H, Shiue C, Fatyol K, Fahlen S, Hydbring P, Soderberg O, Grummt I, Larsson LG, Wright AP (2005) c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat Cell Biol 7:303–310
Mayer C, Zhao J, Yuan X, Grummt I (2004) mTOR-dependent activation of the transcription factor TIF-IA links rRNA synthesis to nutrient availability. Genes Dev 18(4):423–434. doi:10.1101/gad.285504
Hannan KM, Brandenburger Y, Jenkins A, Sharkey K, Cavanaugh A, Rothblum L, Moss T, Poortinga G, McArthur GA, Pearson RB, Hannan RD (2003) mTOR-dependent regulation of ribosomal gene transcription requires S6K1 and is mediated by phosphorylation of the carboxy-terminal activation domain of the nucleolar transcription factor UBF. Mol Cell Biol 23(23):8862–8877. doi:10.1128/mcb.23.23.8862-8877.2003
Zhai W, Comai L (2000) Repression of RNA polymerase I transcription by the tumor suppressor p53. Mol Cell Biol 20:5930–5938. doi:10.1128/mcb.20.16.5930-5938.2000
Budde A, Grummt I (1999) p53 represses ribosomal gene expression. Oncogene 19:1119–1124
Rieker C, Engblom D, Kreiner G, Domanskyi A, Schober A, Stotz S, Neumann M, Yuan X, Grummt I, Schütz G, Parlato R (2011) Nucleolar disruption in dopaminergic neurons leads to oxidative damage and parkinsonism through repression of mammalian target of rapamycin signaling. J Neurosci 31(2):453–460. doi:10.1523/jneurosci.0590-10.2011
Sugimoto M, Kuo ML, Roussel MF, Sherr CJ (2003) Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell 11:415–424. doi:10.1016/S1097-2765(03)00057-1
Bertwistle D, Sugimoto M, Sherr CJ (2004) Physical and functional interactions of the Arf tumor suppressor protein with Nucleophosmin/B23. Mol Cell Biol 24(3):985–996. doi:10.1128/mcb.24.3.985-996.2004
Yamaizumi M, Sugano T (1994) U.V.-induced nuclear accumulation of p53 is evoked through DNA damage of actively transcribed genes independent of the cell cycle. Oncogene 9:2775–2784
Ljungman M, Zhang F (1996) Blockage of RNA polymerase as a possible trigger for u.v. light-induced apoptosis. Oncogene 13:823–831
Trask DK, Muller MT (1988) Stabilization of type I topoisomerase-DNA covalent complexes by actinomycin D. Proc Natl Acad Sci USA 85(5):1417–1421
Ashcroft M, Taya Y, Vousden KH (2000) Stress signals utilize multiple pathways to stabilize p53. Mol Cell Biol 20:3224–3233. doi:10.1128/MCB.20.9.3224-3233.2000
Choong ML, Yang H, Lee MA, Lane DP (2009) Specific activation of the p53 pathway by low dose actinomycin D: a new route to p53 based cyclotherapy. Cell Cycle 8(17):2810–2818
Drygin D, Lin A, Bliesath J, Ho CB, O’Brien SE, Proffitt C, Omori M, Haddach M, Schwaebe MK, Siddiqui-Jain A, Streiner N, Quin JE, Sanij E, Bywater MJ, Hannan RD, Ryckman D, Anderes K, Rice WG (2011) Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res 71(4):1418–1430. doi:10.1158/0008-5472.can-10-1728
Perry RP, Kelley DE (1970) Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol 76(2):127–139. doi:10.1002/jcp.1040760202
Bywater Megan J, Poortinga G, Sanij E, Hein N, Peck A, Cullinane C, Wall M, Cluse L, Drygin D, Anderes K, Huser N, Proffitt C, Bliesath J, Haddach M, Schwaebe Michael K, Ryckman David M, Rice William G, Schmitt C, Lowe Scott W, Johnstone Ricky W, Pearson Richard B, McArthur Grant A, Hannan Ross D (2012) Inhibition of RNA polymerase I as a therapeutic strategy to promote cancer-specific activation of p53. Cancer Cell 22(1):51–65. doi:10.1016/j.ccr.2012.05.019
Vassilev LT (2007) MDM2 inhibitors for cancer therapy. Trends Mol Med 13(1):23–31. doi:10.1016/j.molmed.2006.11.002
Sherr CJ, Weber JD (2000) The ARF/p53 pathway. Curr Opin Genet Dev 10:94–99. doi:10.1016/S0959-437X(99)00038-6
Roth J, Dobbelstein M, Freedman DA, Shenk T, Levine AJ (1998) Nucleo-cytoplasmic shuttling of the hdm2 oncoprotein regulates the levels of the p53 protein via a pathway used by the human immunodeficiency virus rev protein. EMBO J 17:554–564. doi:10.1093/emboj/17.2.554
Hiscox JA (2007) RNA viruses: hijacking the dynamic nucleolus. Nat Rev Microbiol 5:119–127. doi:10.1038/nrmicro1597
Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997) CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060. doi:10.1016/S0092-8674(00)80371-2
Freedman DA, Levine AJ (1998) Nuclear export is required for degradation of endogenous p53 by MDM2 and human papillomavirus E6. Mol Cell Biol 18:7288–7293
Laín S, Midgley C, Sparks A, Lane EB, Lane DP (1999) An inhibitor of nuclear export activates the p53 response and induces the localization of HDM2 and p53 to U1A-positive nuclear bodies associated with the PODs. Exp Cell Res 248:457–472. doi:10.1006/excr.1999.4433
Xirodimas DP, Stephen CW, Lane DP (2001) Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res 270:66–77. doi:10.1006/excr.2001.5314
Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W (2003) Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science 302:1972–1975. doi:10.1126/science.1091362
Boyd MT, Vlatković N, Rubbi CP (2011) The nucleolus directly regulates p53 export and degradation. J Cell Biol 194(5):689–703. doi:10.1083/jcb.201105143
Shirangi TR, Zaika A, Moll UM (2002) Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J 16:420–422. doi:10.1096/fj.01-0617fje
Joseph TW, Zaika A, Moll UM (2003) Nuclear and cytoplasmic degradation of endogenous p53 and HDM2 occurs during down-regulation of the p53 response after multiple types of DNA damage. FASEB J 17:1622–1630. doi:10.1096/fj.02-0931com
Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D (1999) Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1:20–26. doi:10.1038/8991
Zhang Y, Xiong Y (1999) Mutations in the human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol Cell 3:579–591. doi:10.1016/S1097-2765(00)80351-2
Tao W, Levine AJ (1999) p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci USA 96:6937–6941. doi:10.1073/pnas.96.12.6937
Mekhail K, Gunaratnam L, Bonicalzi ME, Lee S (2004) HIF activation by pH-dependent nucleolar sequestration of VHL. Nat Cell Biol 6:642–647. doi:10.1038/ncb1144
Tao W, Levine AJ (1999) Nucleocytoplasmic shuttling of oncoprotein Hdm2 is required for Hdm2-mediated degradation of p53. Proc Natl Acad Sci USA 96:3077–3080. doi:10.1073/pnas.96.6.3077
Stommel JM, Marchenko ND, Jimenez GS, Moll UM, Hope TJ, Wahl GM (1999) A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J 18:1660–1672. doi:10.1093/emboj/18.6.1660
Boyd SD, Tsai KY, Jacks T (2000) An intact HDM2 RING-finger domain is required for nuclear exclusion of p53. Nat Cell Biol 2:563–568. doi:10.1038/35023500
Geyer RK, Yu ZK, Maki CG (2000) The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat Cell Biol 2:569–573. doi:10.1038/35023507
Gu J, Nie L, Wiederschain D, Yuan ZM (2001) Identification of p53 sequence elements that are required for MDM2-mediated nuclear export. Mol Cell Biol 21:8533–8546. doi:10.1128/MCB.21.24.8533-8546.2001
Llanos S, Clark PA, Rowe J, Peters G (2001) Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol 3:445–452. doi:10.1038/35074506
Xirodimas D, Saville MK, Edling C, Lane DP, Lain S (2001) Different effects of p14ARF on the levels of ubiquitinated p53 and Mdm2 in vivo. Oncogene 20:4972–4983
Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH (2000) Identification of a cryptic nucleolar-localization signal in MDM2. Nat Cell Biol 2:179–181. doi:10.1038/35004057
Ahmad Y, Boisvert F-M, Gregor P, Cobley A, Lamond AI (2009) NOPdb: nucleolar Proteome Database—2008 update. Nucleic Acids Res 37(suppl 1):D181–D184. doi:10.1093/nar/gkn804
Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W (2002) Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416:648–653
Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B (2004) Disruption of HAUSP gene stabilizes p53. Nature 428:1 p following 486
Cummins JM, Vogelstein B (2004) HAUSP is required for p53 destabilization. Cell Cycle 3:689–692
Li M, Brooks CL, Kon N, Gu W (2004) A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell 13:879–886
Gray LJ, Bjelogrlic P, Appleyard VCL, Thompson AM, Jolly CE, Lain S, Herrington CS (2007) Selective induction of apoptosis by leptomycin B in keratinocytes expressing HPV oncogenes. Int J Cancer 120(11):2317–2324. doi:10.1002/ijc.22591
Mekhail K, Khacho M, Carrigan A, Hache RR, Gunaratnam L, Lee S (2005) Regulation of ubiquitin ligase dynamics by the nucleolus. J Cell Biol 170:733–744. doi:10.1083/jcb.200506030
Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, Bertrand E (2004) PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16:777–787. doi:10.1016/j.molcel.2004.11.013
Sleeman J (2007) A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci 120(9):1540–1550. doi:10.1242/jcs.001529
Sleeman JE, Ajuh P, Lamond AI (2001) snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 114(24):4407–4419
Pradet-Balade B, Girard C, Boulon S, Paul C, Azzag K, Bordonne R, Bertrand E, Verheggen C (2011) CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J 30(11):2205–2218. doi:10.1038/emboj.2011.128
Muro E, Hoang TQ, Jobart-malfait A, Dl Hernandez-verdun (2008) In nucleoli, the steady state of nucleolar proteins is leptomycin B-sensitive. Biol Cell 100(5):303–313. doi:10.1042/bc20070117
Rubbi CP, Milner J (2000) Non-activated p53 co-localizes with sites of transcription within both the nucleoplasm and the nucleolus. Oncogene 19:85–96
Klibanov SA, O’Hagan HM, Ljungman M (2001) Accumulation of soluble and nucleolar-associated p53 proteins following cellular stress. J Cell Sci 114:1867–1873
Mélèse T, Xue Z (1995) The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol 7(3):319–324. doi:10.1016/0955-0674(95)80085-9
Macias E, Jin AW, Deisenroth C, Bhat K, Mao H, Lindstrom MS, Zhang YP (2010) An ARF-Independent c-MYC-activated tumor suppression pathway mediated by ribosomal protein-Mdm2 interaction. Cancer Cell 18(3):231–243. doi:10.1016/j.ccr.2010.08.007
Polanski R (2009) Identification of MDM2-interacting proteins in renal cell carcinoma and characterization of phenotypic properties acquired by renal cell carcinoma cells which over-express MDM2 and p53. PhD Thesis, Liverpool, Liverpool
Zhang Y, Lu H (2009) Signaling to p53: ribosomal proteins find their way. Cancer Cell 16(5):369–377. doi:10.1016/j.ccr.2009.09.024
Borer RA, Lehner CF, Eppenberger HM, Nigg EA (1989) Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell 56:379–390. doi:10.1016/0092-8674(89)90241-9
Chen D, Huang S (2001) Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 153:169–176. doi:10.1083/jcb.153.1.169
Finch RA, Chan PK (1996) ATP depletion affects NPM translocation and exportation of rRNA from nuclei. Biochem Biophys Res Commun 222:553–558. doi:10.1006/bbrc.1996.0782
Tsai RY, McKay RD (2005) A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol 168:179–184. doi:10.1083/jcb.200409053
Lam YW, Lamond AI, Mann M, Andersen JS (2007) Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17(9):749–760. doi:10.1016/j.cub.2007.03.064
Cassio D (2006) Viable hybrids between adherent cells: generation, yield improvement, and analysis. In: Julio EC (ed) Cell biology. 3rd edn. Academic Press, Burlington, pp 231–235. doi:10.1016/b978-012164730-8/50030-7
Komarova EA, Zelnick CR, Chin D, Zeremski M, Gleiberman AS, Bacus SS, Gudkov AV (1997) Intracellular localization of p53 tumor suppressor protein in gamma-irradiated cells is cell cycle regulated and determined by the nucleus. Cancer Res 57:5217–5220
Marchenko ND, Wolff S, Erster S, Becker K, Moll UM (2007) Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 26:923–934. doi:10.1038/sj.emboj.7601560
Krüger T, Scheer U (2010) p53 localizes to intranucleolar regions distinct from the ribosome production compartments. J Cell Sci 123(8):1203–1208. doi:10.1242/jcs.062398
Maki CG, Howley PM (1997) Ubiquitination of p53 and p21 is differentially affected by ionizing and UV radiation. Mol Cell Biol 17(1):355–363
O’Hagan HM, Ljungman M (2004) Nuclear accumulation of p53 following inhibition of transcription is not due to diminished levels of MDM2. Oncogene 23:5505–5512. doi:10.1038/sj.onc.1207709
Solovei I, Schermelleh L, Albiez H, Cremer T (2006) Detection of cell cycle stages in situ in growing cell populations. In: Celis JE (ed) Cell biology, 3rd edn. Academic Press, Burlington, pp 291–299
Latonen L, Moore HM, Bai B, Jaamaa S, Laiho M (2011) Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 30:790–805. doi:10.1038/onc.2010.469
Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, Kozak K, Csucs G, Lund E, Kutay U (2010) A protein inventory of human ribosome biogenesis reveals an essential function of Exportin 5 in 60S subunit export. PLoS Biol 8(10):e1000522. doi:10.1371/journal.pbio.1000522
Scharf A, Rockel T, von Mikecz A (2007) Localization of proteasomes and proteasomal proteolysis in the mammalian interphase cell nucleus by systematic application of immunocytochemistry. Histochem Cell Biol 127(6):591–601. doi:10.1007/s00418-006-0266-2
Lippincott-Schwartz J, Altan-Bonnet N, Patterson GH (2003) Photobleaching and photoactivation: following protein dynamics in living cells. Nat Cell Biol Suppl:S7–14. doi:10.1038/ncb1032
Karni-Schmidt O, Friedler A, Zupnick A, McKinney K, Mattia M, Beckerman R, Bouvet P, Sheetz M, Fersht A, Prives C (2007) Energy-dependent nucleolar localization of p53 in vitro requires two discrete regions within the p53 carboxyl terminus. Oncogene. doi:10.1038/sj.onc.1210162
Brangwynne CP, Mitchison TJ, Hyman AA (2011) Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci USA 108(11):4334–4339. doi:10.1073/pnas.1017150108
Audas Timothy E, Jacob Mathieu D, Lee S (2012) Immobilization of proteins in the nucleolus by ribosomal intergenic spacer noncoding RNA. Mol Cell 45(2):147–157
Audas TE, Jacob MD, Lee S (2012) The nucleolar detention pathway: a cellular strategy for regulating molecular networks. Cell Cycle 11(11):2059–2062
Moore HM, Bai B, Boisvert F-M, Latonen L, Rantanen V, Simpson JC, Pepperkok R, Lamond AI, Laiho M (2011) Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics 10(10). doi: 10.1074/mcp.M111.009241
Rockel TD, Stuhlmann D, von Mikecz A (2005) Proteasomes degrade proteins in focal subdomains of the human cell nucleus. J Cell Sci 118(22):5231–5242. doi:10.1242/jcs.02642
Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI (2002) Directed proteomic analysis of the human nucleolus. Curr Biol 12:1–11. doi:10.1016/S0960-9822(01)00650-9
Baserga R, Huang C-H, Rossini M, Chang H, Ming P-ML (1976) The role of nuclei and nucleoli in the control of cell proliferation. Cancer Res 36((11 Part 2)):4297–4300
Puvion-Dutilleul F, Macieira-Coelho A (1983) Aging-dependent nucleolar and chromatin changes in cultivated fibroblasts. Cell Biol Int Rep 7(1):61–71. doi:10.1016/0309-1651(83)90105-4
Bowman PD, Meek RL, Daniel CW (1976) Decreased synthesis of nucleolar RNA in aging human cells in vitro. Exp Cell Res 101(2):434–437. doi:10.1016/0014-4827(76)90399-2
Reich NC, Levine AJ (1984) Growth regulation of a cellular tumour antigen, p53, in nontransformed cells. Nature 308(5955):199–201. doi:10.1038/308199a0
Lubbert M, Miller CW, Kahan J, Koeffler HP (1989) Expression, methylation and chromatin structure of the p53 gene in untransformed and human T-cell leukemia virus type I-transformed human T-lymphocytes. Oncogene 4(5):643–651
Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3:577–587. doi:10.1016/S1535-6108(03)00134-X
Zhang Y, Wolf GW, Bhat K, Jin A, Allio T, Burkhart WA, Xiong Y (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol Cell Biol 23:8902–8912. doi:10.1128/MCB.23.23.8902-8912.2003
Gilkes DM, Chen L, Chen J (2006) MDMX regulation of p53 response to ribosomal stress. EMBO J 25:5614–5625. doi:10.1038/sj.emboj.7601424
Pan Y, Chen J (2003) MDM2 promotes ubiquitination and degradation of MDMX. Mol Cell Biol 23(15):5113–5121. doi:10.1128/mcb.23.15.5113-5121.2003
Welf ES, Ahmed S, Johnson HE, Melvin AT, Haugh JM (2012) Migrating fibroblasts reorient directionality by a metastable, PI3 K-dependent mechanism. J Cell Biol 197(1):105–114. doi:10.1083/jcb.201108152
Okamoto K, Taya Y, Nakagama H (2009) Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett 583(17):2710–2714. doi:10.1016/j.febslet.2009.07.021
Dai M-S, Shi D, Jin Y, Sun X–X, Zhang Y, Grossman SR, Lu H (2006) Regulation of the MDM2-p53 pathway by ribosomal protein L11 involves a post-ubiquitination mechanism. J Biol Chem 281(34):24304–24313. doi:10.1074/jbc.M602596200
Sun X–X, Dai M-S, Lu H (2007) 5-Fluorouracil activation of p53 involves an MDM2-ribosomal protein interaction. J Biol Chem 282(11):8052–8059. doi:10.1074/jbc.M610621200
Fumagalli S, Di Cara A, Neb-Gulati A, Natt F, Schwemberger S, Hall J, Babcock GF, Bernardi R, Pandolfi PP, Thomas G (2009) Absence of nucleolar disruption after impairment of 40S ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat Cell Biol 11(4):501–508. doi:10.1038/ncb1858
Granetto C, Ottaggio L, Abbondandolo A, Bonatti S (1996) p53 accumulates in micronuclei after treatment with a DNA breaking chemical, methylnitrosourea, and with the spindle poison, vinblastine. Mutat Res 352:61–64. doi:10.1016/0027-5107(95)00235-9
Fontoura BM, Sorokina EA, David E, Carroll RB (1992) p53 is covalently linked to 5.8S rRNA. Mol Cell Biol 12:5145–5151. doi:10.1038/ncb1858
Fontoura BM, Atienza CA, Sorokina EA, Morimoto T, Carroll RB (1997) Cytoplasmic p53 polypeptide is associated with ribosomes. Mol Cell Biol 17:3146–3154
Chen J, Guo K, Kastan MB (2012) Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem 287(20):16467–16476. doi:10.1074/jbc.M112.349274
Takagi M, Absalon MJ, McLure KG, Kastan MB (2005) Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell 123:49–63. doi:10.1016/j.cell.2005.07.034
Ofir-Rosenfeld Y, Boggs K, Michael D, Kastan MB, Oren M (2008) Mdm2 regulates p53 mRNA translation through inhibitory interactions with ribosomal protein L26. Mol Cell 32(2):180–189. doi:10.1016/j.molcel.2008.08.031
Fu L, Benchimol S (1997) Participation of the human p53 3′UTR in translational repression and activation following gamma-irradiation. EMBO J 16:4117–4125. doi:10.1093/emboj/16.13.4117
Fu L, Minden MD, Benchimol S (1996) Translational regulation of human p53 gene expression. EMBO J 15(16):4392–4401
Maclaine NJ, Hupp TR (2011) How phosphorylation controls p53. Cell Cycle 10(6):916–921. doi:10.4161/cc.10.6.15076
Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H (2004) Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene 23(41):6845–6853. doi:10.1038/sj.onc.1207860
Uetake Y, Sluder G (2010) Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol 20(18):1666–1671. doi:10.1016/j.cub.2010.08.018
Sablina AA, Ilyinskaya GV, Rubtsova SN, Agapova LS, Chumakov PM, Kopnin BP (1998) Activation of p53-mediated cell cycle checkpoint in response to micronuclei formation. J Cell Sci 111(Pt 7):977–984
Hernandez-Verdun D, Robert-Nicoud M, Geraud G, Masson C (1991) Behaviour of nucleolar proteins in nuclei lacking ribosomal genes. A study by confocal laser scanning microscopy. J Cell Sci 98((Pt 1)):99–105
Leung AKL, Gerlich D, Miller G, Lyon C, Lam YW, Lleres D, Daigle N, Zomerdijk J, Ellenberg J, Lamond AI (2004) Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J Cell Biol 166(6):787–800. doi:10.1083/jcb.200405013
Huang H-C, Shi J, Orth JD, Mitchison TJ (2009) Evidence that mitotic exit is a better cancer therapeutic target than spindle assembly. Cancer Cell 16(4):347–358. doi:10.1016/j.ccr.2009.08.020
Raška I, Koberna K, Malínský J, Fidlerová H, Mašata M (2004) The nucleolus and transcription of ribosomal genes. Biol Cell 96(8):579–594. doi:10.1016/j.biolcel.2004.04.015
Longley DB, Harkin DP, Johnston PG (2003) 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer 3(5):330–338. doi:10.1038/nrc1074
Pritchard DM, Watson AJM, Potten CS, Jackman AL, Hickman JA (1997) Inhibition by uridine but not thymidine of p53-dependent intestinal apoptosis initiated by 5-fluorouracil: evidence for the involvement of RNA perturbation. Proc Natl Acad Sci USA 94:1795–1799
Ghoshal K, Jacob ST (1994) Specific inhibition of pre-ribosomal RNA processing in extracts from the lymphosarcoma cells treated with 5-fluorouracil. Cancer Res 54:632–636
Ghoshal K, Jacob ST (1997) An alternative molecular mechanism of action of 5-fluorouracil, a potent anticancer drug. Biochem Pharmacol 53:1569–1575. doi:10.1016/S0006-2952(97)00040-3
Gelasco A, Lippard SJ (1998) NMR solution structure of a DNA dodecamer suplex containing a cis-Diammineplatinum(II) d(GpG) intrastrand cross-link, the major adduct of the anticancer drug cisplatin. Biochemistry (Mosc) 37(26):9230–9239. doi:10.1021/bi973176v
Jordan P, Carmo-Fonseca M (2000) Molecular mechanisms involved in cisplatin cytotoxicity. Cell Mol Life Sci 57(8–9):1229–1235. doi:10.1007/PL00000762
Ljungman M, Lane DP (2004) Transcription—guarding the genome by sensing DNA damage. Nat Rev Cancer 4:727–737. doi:10.1038/nrc1435
Ohndorf U-M, Rould MA, He Q, Pabo CO, Lippard SJ (1999) Basis for recognition of cisplatin-modified DNA by high-mobility-group proteins. Nature 399:708–712. doi:10.1038/21460
Jordan P, Carmo-Fonseca M (1998) Cisplatin inhibits synthesis of ribosomal RNA in vivo. Nucleic Acids Res 26:2831–2836. doi:10.1093/nar/26.12.2831
Zhai X, Beckmann H, Jantzen HM, Essigmann JM (1998) Cisplatin-DNA adducts inhibit ribosomal RNA synthesis by hijacking the transcription factor human upstream binding factor. Biochemistry (Mosc) 37:16307–16315. doi:10.1021/bi981708h
Huang R, Wu T, Xu L, Liu A, Ji Y, Hu G (2002) Upstream binding factor up-regulated in hepatocellular carcinoma is related to the survival and cisplatin-sensitivity of cancer cells. FASEB J 16:293–301. doi:10.1096/fj.01-0687com
Rabik CA, Dolan ME (2007) Molecular mechanisms of resistance and toxicity associated with platinating agents. Cancer Treat Rev 33(1):9–23. doi:10.1016/j.ctrv.2006.09.006
McKeage MJ, Hsu T, Screnci D, Haddad G, Baguley BC (2001) Nucleolar damage correlates with neurotoxicity induced by different platinum drugs. Br J Cancer 85:1219–1225. doi:10.1054/bjoc.2001.2024
Pizzolato JF, Saltz LB (2003) The camptothecins. The Lancet 361(9376):2235–2242. doi:10.1016/s0140-6736(03)13780-4
Mao Y, Mehl IR, Muller MT (2002) Subnuclear distribution of topoisomerase I is linked to ongoing transcription and p53 status. Proc Natl Acad Sci USA 99:1235–1240. doi:10.1073/pnas.022631899
Buckwalter CA, Lin AH, Tanizawa A, Pommier YG, Cheng Y-C, Kaufmann SH (1996) RNA synthesis inhibitors alter the subnuclear distribution of DNA topoisomerase I. Cancer Res 56:1674–1681
Morris EJ, Geller HM (1996) Induction of neuronal apoptosis by camptothecin, an inhibitor of DNA topoisomerase-I: evidence for cell cycle-independent toxicity. J Cell Biol 134:757–770. doi:10.1083/jcb.134.3.757
Kalita K, Makonchuk D, Gomes C, Zheng J–J, Hetman M (2008) Inhibition of nucleolar transcription as a trigger for neuronal apoptosis. J Neurochem 105(6):2286–2299. doi:10.1111/j.1471-4159.2008.05316.x
Tomkins CE, Edwards SN, Tolkovsky AM (1994) Apoptosis is induced in post-mitotic rat sympathetic neurons by arabinosides and topoisomerase II inhibitors in the presence of NGF. J Cell Sci 107:1499–1507
Drygin D, Siddiqui-Jain A, O’Brien S, Schwaebe M, Lin A, Bliesath J, Ho CB, Proffitt C, Trent K, Whitten JP, Lim JKC, Von Hoff D, Anderes K, Rice WG (2009) Anticancer activity of CX-3543: a direct inhibitor of rRNA biogenesis. Cancer Res 69(19):7653–7661. doi:10.1158/0008-5472.can-09-1304
Zhu Y, Poyurovsky MV, Li YC, Biderman L, Stahl J, Jacq X, Prives C (2009) Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol Cell 35(3):316–326. doi:10.1016/j.molcel.2009.07.014
Balasubramanian S, Hurley LH, Neidle S (2011) Targeting G-quadruplexes in gene promoters: a novel anticancer strategy? Nat Rev Drug Discov 10(4):261–275. doi:10.1038/nrd3428
Latonen L, Kurki S, Pitkänen K, Laiho M (2003) p53 and MDM2 are regulated by PI-3-kinases on multiple levels under stress induced by UV radiation and proteasome dysfunction. Cell Signal 15(1):95–102. doi:10.1016/s0898-6568(02)00044-x
Acknowledgments
Work in our laboratory was supported by Mersey Kidney Research, Cancer Research UK, The North West Cancer Research Fund, The Isle of Man Anti Cancer Association, and by The Hampson Trust of the Royal Liverpool University Hospital.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vlatković, N., Boyd, M.T. & Rubbi, C.P. Nucleolar control of p53: a cellular Achilles’ heel and a target for cancer therapy. Cell. Mol. Life Sci. 71, 771–791 (2014). https://doi.org/10.1007/s00018-013-1361-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1361-x