Abstract
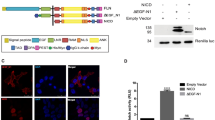
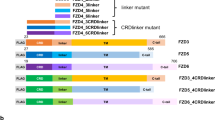
The Notch signalling pathway has a crucial function in determining cell fates in multiple tissues within metazoan organisms1. On binding to ligands, the Notch receptor is cleaved proteolytically and releases its intracellular domain (NotchICD). The NotchICD enters the nucleus and acts cooperatively with other factors to stimulate the transcription of target genes. High levels of Notch-mediated transcriptional activation require the formation of a ternary complex consisting of NotchICD, CSL (CBF-1, suppressor of hairless, LAG-1) and a Mastermind family member2,3,4,5. However, it is still not clear how the formation of the ternary complex is regulated. Here we show that Nemo-like kinase (NLK) negatively regulates Notch-dependent transcriptional activation by decreasing the formation of this ternary complex. Using a biochemical screen, we identified Notch as a new substrate of NLK. NLK-phosphorylated Notch1ICD is impaired in its ability to form a transcriptionally active ternary complex. Furthermore, knockdown of NLK leads to hyperactivation of Notch signalling and consequently decreases neurogenesis in zebrafish. Our results both define a new function for NLK and reveal a previously unidentified mode of regulation in the Notch signalling pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Artavanis-Tsakonas, S., Rand, M. D. & Lake, R. J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Bray, S. J. Notch signalling: a simple pathway becomes complex. Nature Rev. Mol. Cell Biol. 7, 678–689 (2006).
Wu, L. et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nature Genet. 26, 484–489 (2000).
Kitagawa, M. et al. A human protein with sequence similarity to Drosophila mastermind coordinates the nuclear form of notch and a CSL protein to build a transcriptional activator complex on target promoters. Mol. Cell. Biol. 21, 4337–4346 (2001).
Oyama, T. et al. Mastermind-1 is required for Notch signal-dependent steps in lymphocyte development in vivo. Proc. Natl Acad. Sci. USA 104, 9764–9769 (2007).
Choi, K. W. & Benzer, S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell 78, 125–136 (1994).
Meneghini, M. D. et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature 399, 793–797 (1999).
Thorpe, C. J. & Moon, R. T. nemo-like kinase is an essential co-activator of Wnt signaling during early zebrafish development. Development 131, 2899–2909 (2004).
Kortenjann, M. et al. Abnormal bone marrow stroma in mice deficient for nemo-like kinase, Nlk. Eur. J. Immunol. 31, 3580–3587 (2001).
Ishitani, T. et al. The TAK1–NLK–MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature 399, 798–802 (1999).
Ishitani, T., Ninomiya-Tsuji, J. & Matsumoto, K. Regulation of lymphoid enhancer factor 1/T-cell factor by mitogen-activated protein kinase-related Nemo-like kinase-dependent phosphorylation in Wnt/β-catenin signaling. Mol. Cell. Biol. 23, 1379–1389 (2003).
Kanei-Ishii, C. et al. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 18, 816–829 (2004).
Kojima, H. et al. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFβ-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl Acad. Sci. USA 102, 4524–4529 (2005).
Ohkawara, B. et al. Role of the TAK1–NLK–STAT3 pathway in TGF-β-mediated mesoderm induction. Genes Dev. 18, 381–6 (2004).
Verheyen, E. M., Purcell, K. J., Fortini, M. E. & Artavanis-Tsakonas, S. Analysis of dominant enhancers and suppressors of activated Notch in Drosophila. Genetics 144, 1127–1141 (1996).
Kankel, M. W. et al. Investigating the genetic circuitry of mastermind in Drosophila, a notch signal effector. Genetics 177, 2493–2505 (2007).
Zhou, S. et al. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol. Cell. Biol. 20, 2400–2410 (2000).
Takke, C., Dornseifer, P., v. Weizsacker, E. & Campos-Ortega, J. A. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development 126, 1811–1821 (1999).
Yeo, S., Kim, M. J., Kim, H., Huh, T. & Chitnis, A. B. Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 301, 555–567 (2007).
Hyodo-Miura, J. et al. Involvement of NLK and Sox11 in neural induction in Xenopus development. Genes Cells 7, 487–496 (2002).
Bigas, A., Martin, D. I. & Milner, L. A. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol. Cell Biol. 18, 2324–2333 (1998).
Inglés-Esteve, J., Espinosa, L., Milner, L. A., Caelles, C. & Bigas, A. Phosphorylation of Ser2078 modulates the Notch2 function in 32D cell differentiation. J. Biol. Chem. 276, 44873–44880 (2001).
Kurooka, H., Kuroda, K. & Honjo, T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 26, 5448–5455 (1998).
Kurooka, H. & Honjo, T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 275, 17211–17220 (2000).
Pufall, M. A. et al. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science 309, 142–145 (2005).
Zhao, Y., Tong, C. & Jiang, J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature 450, 252–258 (2007).
Minoguchi, S. et al. RPB-L, a transcription factor related to RBP-Jκ. Mol. Cell. Biol. 17, 2679–2687 (1997).
Raya, A. et al. Notch activity induces Nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes Dev. 17, 1213–1218 (2003).
Kim, C. H. et al. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci. Lett. 216, 109–112 (1996).
Mizutani, T. et al. Conservation of the biochemical mechanisms of signal transduction among mammalian Notch family members. Proc. Natl Acad. Sci. USA 98, 9026–9031 (2001).
Shimizu, K. et al. Functional diversity among Notch1, Notch2, and Notch3 receptors. Biochem. Biophys. Res. Commun. 291, 775–779 (2002).
Lin, S. E. et al. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J. Biol. Chem. 277, 50612–50620 (2002).
Wettstein, D. A., Turner, D. L. & Kintner, C. The Xenopus homolog of Drosphila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development 124, 693–702 (1997).
Acknowledgements
We thank T. Honjo, T. C. Südhof, U. Lendahl, K. Yasutomo, C. Kintner, D. Hayward, J. D. Griffin, L. Wu, M. Takeichi, S. Chiba, A. Kikuchi, H. Fujisawa and K. Hozumi for providing plasmid vectors, antibody and cultured cells; H. Matsuo for technical assistance; members of the H. Aiba laboratory (especially T. Sunohara) for technical advice; and J. Ninomiya-Tsuji for helpful discussions. This research was supported by the Yamada Science Foundation (T.I.), the Astellas Foundation for Research on Metabolic Disorders (T.I. and M.I.), the Program for Improvement of Research Environment for Young Researchers from SCF commissioned by MEXT of Japan (T.I. and M.I.), and the Grants-in-Aid for Scientific Research programs in Japan (T.I., K.M. and M.I.).
Author information
Authors and Affiliations
Contributions
T.I. designed the research, performed most of the experiments, analysed data and wrote the paper. T.H., M.S., M. Isoda and S.I. performed experiments and analysed data. K.H. participated in discussions and helped write the paper. M.K. designed the research and performed the experiments, analysed data and wrote the paper. K.M. designed the initial experiments to identify NLK substrates, participated discussions, and helped write the paper. M. Itoh designed the project and experiments, supervised the research, coordinated experiments and wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1585 kb)
Rights and permissions
About this article
Cite this article
Ishitani, T., Hirao, T., Suzuki, M. et al. Nemo-like kinase suppresses Notch signalling by interfering with formation of the Notch active transcriptional complex. Nat Cell Biol 12, 278–285 (2010). https://doi.org/10.1038/ncb2028
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2028
This article is cited by
-
Phosphorylation of MAVS/VISA by Nemo-like kinase (NLK) for degradation regulates the antiviral innate immune response
Nature Communications (2019)
-
Src kinase phosphorylates Notch1 to inhibit MAML binding
Scientific Reports (2018)
-
Matrin-3 is essential for fibroblast growth factor 2-dependent maintenance of neural stem cells
Scientific Reports (2018)
-
C-terminal deletion of NOTCH1 intracellular domain (N1ICD) increases its stability but does not amplify and recapitulate N1ICD-dependent signalling
Scientific Reports (2017)
-
Phosphorylation of Suppressor of Hairless impedes its DNA-binding activity
Scientific Reports (2017)