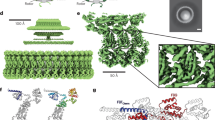
Abstract
The flagellar motor drives the rotation of flagellar filaments at hundreds of revolutions per second1,2, efficiently propelling bacteria through viscous media3. The motor uses the potential energy from an electrochemical gradient of cations4,5 across the cytoplasmic membrane to generate torque. A rapid switch from anticlockwise to clockwise rotation determines whether a bacterium runs smoothly forward or tumbles to change its trajectory6,7. A protein called FliG forms a ring in the rotor of the flagellar motor that is involved in the generation of torque8,9,10,11,12,13 through an interaction with the cation-channel-forming stator subunit MotA12. FliG has been suggested to adopt distinct conformations that induce switching but these structural changes and the molecular mechanism of switching are unknown. Here we report the molecular structure of the full-length FliG protein, identify conformational changes that are involved in rotational switching and uncover the structural basis for the formation of the FliG torque ring. This allows us to propose a model of the complete ring and switching mechanism in which conformational changes in FliG reverse the electrostatic charges involved in torque generation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
19 August 2010
Three mistakes were corrected for the print issue.
References
Lowe, G., Meister, M. & Berg, H. C. Rapid rotation of flagellar bundles in swimming bacteria. Nature 325, 637–640 (1987)
Magariyama, Y. et al. Very fast flagellar rotation. Nature 371, 752 (1994)
Berg, H. C. & Anderson, R. A. Bacteria swim by rotating their flagellar filaments. Nature 245, 380–382 (1973)
Manson, M. D., Tedesco, P., Berg, H. C., Harold, F. M. & Van der Drift, C. A protonmotive force drives bacterial flagella. Proc. Natl Acad. Sci. USA 74, 3060–3064 (1977)
Hirota, N. & Imae, Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J. Biol. Chem. 258, 10577–10581 (1983)
Berg, H. C. & Brown, D. A. Chemotaxis in Escherichia coli analysed by three-dimensional tracking. Nature 239, 500–504 (1972)
Turner, L., Ryu, W. S. & Berg, H. C. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182, 2793–2801 (2000)
Irikura, V. M., Kihara, M., Yamaguchi, S., Sockett, H. & Macnab, R. M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175, 802–810 (1993)
Lloyd, S. A. & Blair, D. F. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J. Mol. Biol. 266, 733–744 (1997)
Lloyd, S. A., Tang, H., Wang, X., Billings, S. & Blair, D. F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J. Bacteriol. 178, 223–231 (1996)
Yorimitsu, T., Mimaki, A., Yakushi, T. & Homma, M. The conserved charged residues of the C-terminal region of FliG, a rotor component of the Na+-driven flagellar motor. J. Mol. Biol. 334, 567–583 (2003)
Zhou, J., Lloyd, S. A. & Blair, D. F. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl Acad. Sci. USA 95, 6436–6441 (1998)
Lloyd, S. A., Whitby, F. G., Blair, D. F. & Hill, C. P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400, 472–475 (1999)
Brown, P. N., Hill, C. P. & Blair, D. F. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21, 3225–3234 (2002)
Francis, N. R., Irikura, V. M., Yamaguchi, S., DeRosier, D. J. & Macnab, R. M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc. Natl Acad. Sci. USA 89, 6304–6308 (1992)
Ueno, T., Oosawa, K. & Aizawa, S. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J. Mol. Biol. 227, 672–677 (1992)
Kihara, M., Miller, G. U. & Macnab, R. M. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182, 3022–3028 (2000)
Togashi, F., Yamaguchi, S., Kihara, M., Aizawa, S. I. & Macnab, R. M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179, 2994–3003 (1997)
Van Way, S. M., Millas, S. G., Lee, A. H. & Manson, M. D. Rusty, jammed, and well-oiled hinges: mutations affecting the interdomain region of FliG, a rotor element of the Escherichia coli flagellar motor. J. Bacteriol. 186, 3173–3181 (2004)
Andrade, M. A., Petosa, C., O'Donoghue, S. I., Muller, C. W. & Bork, P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 309, 1–18 (2001)
Conti, E., Uy, M., Leighton, L., Blobel, G. & Kuriyan, J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94, 193–204 (1998)
Huber, A. H., Nelson, W. J. & Weis, W. I. Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90, 871–882 (1997)
Brown, P. N., Terrazas, M., Paul, K. & Blair, D. F. Mutational analysis of the flagellar protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189, 305–312 (2007)
Marykwas, D. L. & Berg, H. C. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J. Bacteriol. 178, 1289–1294 (1996)
Thomas, D. R., Francis, N. R., Xu, C. & DeRosier, D. J. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 7039–7048 (2006)
Duke, T. A., Le Novere, N. & Bray, D. Conformational spread in a ring of proteins: a stochastic approach to allostery. J. Mol. Biol. 308, 541–553 (2001)
Bai, F. et al. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science 327, 685–689 (2010)
Thomas, D., Morgan, D. G. & DeRosier, D. J. Structures of bacterial flagellar motors from two FliF–FliG gene fusion mutants. J. Bacteriol. 183, 6404–6412 (2001)
Liu, J. et al. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C ring assembly flexion. J. Bacteriol. 191, 5026–5036 (2009)
Murphy, G. E., Leadbetter, J. R. & Jensen, G. J. In situ structure of the complete Treponema primitia flagellar motor. Nature 442, 1062–1064 (2006)
Leslie, A. G. W. in Joint CCP4 and ESF-EACMB Newsletter on Protein Cystallography No. 26 (Daresbury Laboratory, 1992)
Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 (1994)
Leahy, D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. Structure of a fibronectin type III domain from tenascin phased by MAD analysis of the selenomethionyl protein. Science 258, 987–991 (1992)
Sheldrick, G. M. A short history of SHELX. Acta Crystallogr. A 64, 112–122 (2008)
Fortelle, E. & Bricogne, G. Maximum-likelihood heavy-atom parameter refinement for the multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 276, 472–494 (1997)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 (2007)
Dolinsky, T. J. et al. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007)
Humphrey, W., Dalke, A. & Schulten, K. VMD: visual molecular dynamics. J. Mol. Graph. 14, 33–38 (1996)
Acknowledgements
We thank the staff at beamline ID14-1 at the ESRF, and at beamlines 14-ID (BioCARS) and 23-ID (General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT)) at the APS for their support. Use of the APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science (contract DE-AC02-06CH11357), and use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources (grant RR007707). GM/CA-CAT was funded in whole or in part with federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). This work was supported by the Australian Synchrotron Research Program of the Australian Nuclear Science Technology Organization. D. Thomas is acknowledged for providing us with the electron microscopy density map of the clockwise-locked S. typhimurium mutant and O. Perisic and H. Huber are acknowledged for cloning vectors (pOPT-GST) and A. aeolicus genomic DNA, respectively. D.S. was initially funded by an MRC Career development award (UK) and C.C. by an MRC predoctoral fellowship (UK).
Author information
Authors and Affiliations
Contributions
L.K.L. collected synchrotron data, solved and analysed the structure, wrote software to generate FliG rings and switch morphs, and wrote the manuscript, M.D. collected synchrotron data, M.D. and M.A.G. purified and crystallized selenomethionine-containing protein, C.C. cloned, purified and crystallized the native protein, D.S. conceived and coordinated project, collected synchrotron data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2, Supplementary Figures 1-11 with legends, full legends for Supplementary Movies 1-6 and References. (PDF 3066 kb)
Supplementary Movie 1
This animated movie shows the formation of the FliG ring. (MOV 30576 kb)
Supplementary Movie 2
This movie shows the Interpolation of the M236 phi and F237 phi angles between the C-terminal conformations of FliGFL (CterFL) and FliGMC (CterMC). (MOV 908 kb)
Supplementary Movie 3
This movie shows a rotating FliG monomer displaying the equivalent residues at the location of all known rotationally biased mutants. (MOV 1526 kb)
Supplementary Movie 4
This movie shows parallels between the ARMC – ARMM+1 right-handed superhelix in the FliG multimer and a eukaryotic ARM superhelix in β-catenin. (MOV 3948 kb)
Supplementary Movie 5
This movie shows the structure of the FliGUNIT. (MOV 978 kb)
Supplementary Movie 6
This movie shows the reversal of the torque generating charges in the FliG ring. (MOV 1952 kb)
Rights and permissions
About this article
Cite this article
Lee, L., Ginsburg, M., Crovace, C. et al. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466, 996–1000 (2010). https://doi.org/10.1038/nature09300
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09300
This article is cited by
-
Structural insights into the mechanism of archaellar rotational switching
Nature Communications (2022)
-
Structures of the stator complex that drives rotation of the bacterial flagellum
Nature Microbiology (2020)
-
A coevolution-guided model for the rotor of the bacterial flagellar motor
Scientific Reports (2018)
-
Rotational direction of flagellar motor from the conformation of FliG middle domain in marine Vibrio
Scientific Reports (2018)
-
Bacterial flagellar switching: a molecular mechanism directed by the logic of an electric motor
Journal of Molecular Modeling (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.