-
PDF
- Split View
-
Views
-
Cite
Cite
Daisuke Hirata, Tsutomu Mano, Alexei V. Abramov, Gennady F. Baryshnikov, Pavel A. Kosintsev, Alexandr A. Vorobiev, Evgeny G. Raichev, Hiroshi Tsunoda, Yayoi Kaneko, Koichi Murata, Daisuke Fukui, Ryuichi Masuda, Molecular Phylogeography of the Brown Bear (Ursus arctos) in Northeastern Asia Based on Analyses of Complete Mitochondrial DNA Sequences, Molecular Biology and Evolution, Volume 30, Issue 7, July 2013, Pages 1644–1652, https://doi.org/10.1093/molbev/mst077
Close - Share Icon Share
Abstract
To further elucidate the migration history of the brown bears (Ursus arctos) on Hokkaido Island, Japan, we analyzed the complete mitochondrial DNA (mtDNA) sequences of 35 brown bears from Hokkaido, the southern Kuril Islands (Etorofu and Kunashiri), Sakhalin Island, and the Eurasian Continent (continental Russia, Bulgaria, and Tibet), and those of four polar bears. Based on these sequences, we reconstructed the maternal phylogeny of the brown bear and estimated divergence times to investigate the timing of brown bear migrations, especially in northeastern Eurasia. Our gene tree showed the mtDNA haplotypes of all 73 brown and polar bears to be divided into eight divergent lineages. The brown bear on Hokkaido was divided into three lineages (central, eastern, and southern). The Sakhalin brown bear grouped with eastern European and western Alaskan brown bears. Etorofu and Kunashiri brown bears were closely related to eastern Hokkaido brown bears and could have diverged from the eastern Hokkaido lineage after formation of the channel between Hokkaido and the southern Kuril Islands. Tibetan brown bears diverged early in the eastern lineage. Southern Hokkaido brown bears were closely related to North American brown bears.
Introduction
The brown bear (Ursus arctos) is widely distributed in the Holarctic region, from Europe to North America. Among the continental islands of northeastern Asia, brown bears occur on Hokkaido Island and the southern Kuril Islands. The phylogeography of the brown bear has been studied in Europe, North America, and Asia. In addition, nomenclature for geographically distinct mitochondrial DNA (mtDNA) clades was proposed by Leonard et al. (2000) and extended by Barnes et al. (2002), Miller et al. (2006), and Calvignac et al. (2008, 2009).
European brown bears are divided into two major lineages, eastern (clade 3a) and western (clade 1), based on mtDNA control region sequences (Taberlet and Bouvet 1994). The eastern lineage is widely distributed on the Eurasian Continent, from northeastern Europe to Far Eastern Russia (Saarma et al. 2007; Korsten et al. 2009; Murtskhvaladze et al. 2010; Tammeleht et al. 2010). The western lineage comprises two groups: the Iberian lineage (clade 1a) and the Balkan/Italian lineage (clade 1b) (Taberlet and Bouvet 1994; Kohn et al. 1995). In North America, four clades have been identified among extant brown bears: clade 2a is distributed in the Admiralty, Baranof, and Chichagof (ABC) island group in southeastern Alaska, clade 3a in western Alaska, clade 3b in eastern Alaska, and clade 4 in continental North America (Talbot and Shields 1996; Waits et al. 1998). mtDNA analyses show the polar bear (Ursus maritimus) to be embedded within the brown bear clade and to be most closely related to the ABC islands brown bear (clade 2a) (Cronin et al. 1991; Shields and Kocher 1991; Talbot and Shields 1996; Shields et al. 2000; Lindqvist et al. 2010; Miller et al. 2012).
Phylogenetic analyses of the mtDNA control region and cytochrome b (cyt b) showed that the brown bear population on Hokkaido Island, Japan, comprises three lineages (central, eastern, and southern; Matsuhashi et al. 1999); these lineages differ in the proportions of the skull (Baryshnikov et al. 2004; Baryshnikov and Puzachenko 2009). Matsuhashi et al. (1999) suggested that the three lineages diverged on the Eurasian Continent prior to dispersal onto Hokkaido via land bridges in different three periods: the southern lineage colonized first, followed by the eastern and then the central lineages. Matsuhashi et al. (2001) found the central Hokkaido lineage to be most closely related to clade 3a containing eastern European and western Alaskan bears, and the eastern Hokkaido lineage to be most closely related to the eastern Alaskan lineage (clade 3b). Although the mtDNA control region phylogeny showed the southern Hokkaido lineage to be most closely related to the Tibetan brown bear, the cyt b phylogeny did not support this relationship. In addition, Miller et al. (2006) found the southern Hokkaido lineage, which they labeled clade 3d, group with the Tibetan brown bear (clade 5). In contrast, Korsten et al. (2009), Davison et al. (2011), and Edwards et al. (2011) found that southern Hokkaido lineage to group with the North American lineage, labeled as clade 4. In summary, there is a possibility that the southern Hokkaido lineage groups with North American lineage and Tibetan brown bears are recognized as a distinct lineage.
Interspecific phylogenies of bears have recently been reconstructed using the complete mitochondrial genome (Yu et al. 2007; Bon et al. 2008; Krause et al. 2008; Lindqvist et al. 2010; Miller et al. 2012). When this approach has applied to intraspecific analyses of bears, complete mtDNA sequences have the potential to greatly facilitate phylogeographic interpretation (Davison et al. 2011). Shorter mtDNA regions (e.g., control region and cyt b) used for phylogenetic analyses may bias time estimates due to homoplasy and are generally associated with considerable uncertainty (Davison et al. 2011).
To further elucidate the migration history of the three lineages of brown bears on Hokkaido Island, it is important to clarify the phylogenetic relationships between the southern Hokkaido and Tibetan brown bears, and among brown bears from Hokkaido and neighboring areas such as the southern Kuril Islands (Etorofu and Kunashiri) and Sakhalin. In this study, we analyzed complete mtDNA sequences from 35 brown bears from Hokkaido, the southern Kuril Islands (Etorofu and Kunashiri), Sakhalin, and the Eurasian Continent (Russia, Bulgaria, and Tibet) to reconstruct maternal phylogeny. We also determined the complete mtDNA sequences from four polar bears. Using Bayesian analysis, we inferred times of divergence of these bear populations to shed light on their migration history.
Results
Phylogenetic Relationships within the Brown Bears
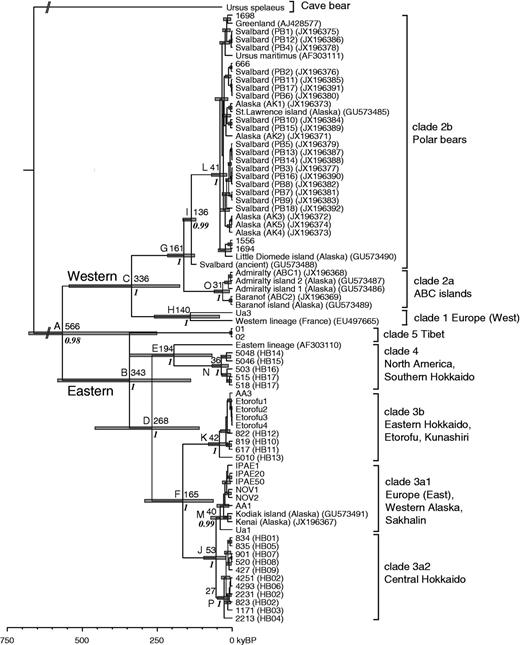
Complete mtDNA sequences for 35 brown bears and four polar bears were successfully determined. The maximum-likelihood (ML) and Bayesian inference (BI) phylogenies were inferred using the newly obtained sequences and 38 previously published complete mtDNA sequences from brown bears and polar bears, with the cave bear as an outgroup. The ML and BI analyses yielded identical tree topologies showing the 73 brown and polar bears divided into eight clades (1, 2a, 2b, 3a1, 3a2, 3b, 4, and 5) (fig. 2; supplementary fig. S1, Supplementary Material online). Brown bears were divided into a western lineage (clades 1, 2a, and 2b) and an eastern lineage (clades 3a1, 3a2, 3b, 4, and 5), with clade 3a further subdivided into clades 3a1 and 3a2 based on five parsimony-informative sites. The polar bears formed clade 2b embedded within the western lineage, which otherwise contained brown bears, and which was the sister clade to the ABC islands group of brown bears (clade 2a).
The brown bears on Hokkaido fell into three clades: 3a2 (central Hokkaido), 3b (eastern Hokkaido), and 4 (southern Hokkaido) (fig. 2; supplementary fig. S1, Supplementary Material online). Clade 3a2 was the sister clade to 3a1, which contained eastern European, western Alaskan, and Sakhalin brown bears. Clade 3b (bootstrap value, 100%; posterior probability, 1.0) (supplementary fig. S1, Supplementary Material online) contained eastern Hokkaido, Etorofu, and Kunashiri brown bears (fig. 2; supplementary fig. S1, Supplementary Material online), with three of four individuals from Etorofu (Etorofu 2, 3, and 4) sharing the same haplotype. Southern Hokkaido bears grouped with North American bears in clade 4 (bootstrap value, 92.8%; posterior probability, 1.0), although the divergence was deep (fig. 2; supplementary fig. S1, Supplementary Material online). Tibetan brown bears, comprising clade 5, were basal in the eastern lineage. Two individuals from the Balkan Mountains in Bulgaria fell into different clades, sample Ua3 into clade 1 in the western lineage and sample Ua1 into clade 3a1 in the eastern lineage.
Divergence Times
Divergence times were estimated by the BI method using internal calibration based on two radiocarbon-dated ancient DNA samples. This permitted the time to the most recent common ancestor (TMRCA) to be estimated for brown bear clades (fig. 2 and table 1). TMRCA estimates are summarized in table 1.
Bayesian Age Estimates (kyBP) for the MRCA of the Brown Bear Clades.
| Node . | MRCA . | Node Age (95% HPD) . | |||
|---|---|---|---|---|---|
| This Studya . | Lindqvist et al. (2010)a . | Davison et al. (2011)b . | Miller et al. (2012)a . | ||
| A | All brown bears | 566 (251–944) | 490 (360–620) | 263 (162–400) | 509 |
| B | Clades 3a, 3b, 4, and 5 | 343 (139–582) | — | — | — |
| C | Clades 1 and 2 | 336 (174–545) | 310 (240–400) | — | 314 |
| D | Clades 3 and 4 | 268 (109–457) | — | 140 (87–213) | — |
| E | Clade 4 | 194 (67–399) | — | 87 (42–147) | — |
| F | Clades 3a and 3b | 165 (63–292) | — | 92 (51–133) | — |
| G | Clade 2 | 161 (125–216) | 152 (131–177) | 160 (124–210) | 162 |
| H | Clade 1 | 140 (42–260) | — | 100 (49–164) | — |
| I | Clade 2b: all polar bears | 136 (120–164) | 134 (122–149) | 146 (120–179) | 136 |
| J | Clade 3a | 53 (21–95) | — | 49 (18–86) | — |
| K | Clade 3b | 42 (14–80) | — | 75 (43–113) | — |
| L | Clade 2b: extant polar bears | 41 (18–68) | 44 | — | 52 |
| M | Clade3a1 | 40 (15–72) | — | — | — |
| N | Clade 4: Southern Hokkaido | 36 (12–67) | — | — | — |
| O | Clade 2a | 31 (9–60) | 28 | 45 (10–91) | — |
| P | Clade 3a2 | 27 (10–49) | — | — | — |
| Node . | MRCA . | Node Age (95% HPD) . | |||
|---|---|---|---|---|---|
| This Studya . | Lindqvist et al. (2010)a . | Davison et al. (2011)b . | Miller et al. (2012)a . | ||
| A | All brown bears | 566 (251–944) | 490 (360–620) | 263 (162–400) | 509 |
| B | Clades 3a, 3b, 4, and 5 | 343 (139–582) | — | — | — |
| C | Clades 1 and 2 | 336 (174–545) | 310 (240–400) | — | 314 |
| D | Clades 3 and 4 | 268 (109–457) | — | 140 (87–213) | — |
| E | Clade 4 | 194 (67–399) | — | 87 (42–147) | — |
| F | Clades 3a and 3b | 165 (63–292) | — | 92 (51–133) | — |
| G | Clade 2 | 161 (125–216) | 152 (131–177) | 160 (124–210) | 162 |
| H | Clade 1 | 140 (42–260) | — | 100 (49–164) | — |
| I | Clade 2b: all polar bears | 136 (120–164) | 134 (122–149) | 146 (120–179) | 136 |
| J | Clade 3a | 53 (21–95) | — | 49 (18–86) | — |
| K | Clade 3b | 42 (14–80) | — | 75 (43–113) | — |
| L | Clade 2b: extant polar bears | 41 (18–68) | 44 | — | 52 |
| M | Clade3a1 | 40 (15–72) | — | — | — |
| N | Clade 4: Southern Hokkaido | 36 (12–67) | — | — | — |
| O | Clade 2a | 31 (9–60) | 28 | 45 (10–91) | — |
| P | Clade 3a2 | 27 (10–49) | — | — | — |
Note.—Nodes refer to those in fig. 2. — indicates that no divergence time estimations are available. Node ages without parentheses have no information on 95% HPD.
aDivergence times were estimated based on the complete mtDNA sequences.
bDivergence times were estimated based on the short mtDNA control region sequences.
Bayesian Age Estimates (kyBP) for the MRCA of the Brown Bear Clades.
| Node . | MRCA . | Node Age (95% HPD) . | |||
|---|---|---|---|---|---|
| This Studya . | Lindqvist et al. (2010)a . | Davison et al. (2011)b . | Miller et al. (2012)a . | ||
| A | All brown bears | 566 (251–944) | 490 (360–620) | 263 (162–400) | 509 |
| B | Clades 3a, 3b, 4, and 5 | 343 (139–582) | — | — | — |
| C | Clades 1 and 2 | 336 (174–545) | 310 (240–400) | — | 314 |
| D | Clades 3 and 4 | 268 (109–457) | — | 140 (87–213) | — |
| E | Clade 4 | 194 (67–399) | — | 87 (42–147) | — |
| F | Clades 3a and 3b | 165 (63–292) | — | 92 (51–133) | — |
| G | Clade 2 | 161 (125–216) | 152 (131–177) | 160 (124–210) | 162 |
| H | Clade 1 | 140 (42–260) | — | 100 (49–164) | — |
| I | Clade 2b: all polar bears | 136 (120–164) | 134 (122–149) | 146 (120–179) | 136 |
| J | Clade 3a | 53 (21–95) | — | 49 (18–86) | — |
| K | Clade 3b | 42 (14–80) | — | 75 (43–113) | — |
| L | Clade 2b: extant polar bears | 41 (18–68) | 44 | — | 52 |
| M | Clade3a1 | 40 (15–72) | — | — | — |
| N | Clade 4: Southern Hokkaido | 36 (12–67) | — | — | — |
| O | Clade 2a | 31 (9–60) | 28 | 45 (10–91) | — |
| P | Clade 3a2 | 27 (10–49) | — | — | — |
| Node . | MRCA . | Node Age (95% HPD) . | |||
|---|---|---|---|---|---|
| This Studya . | Lindqvist et al. (2010)a . | Davison et al. (2011)b . | Miller et al. (2012)a . | ||
| A | All brown bears | 566 (251–944) | 490 (360–620) | 263 (162–400) | 509 |
| B | Clades 3a, 3b, 4, and 5 | 343 (139–582) | — | — | — |
| C | Clades 1 and 2 | 336 (174–545) | 310 (240–400) | — | 314 |
| D | Clades 3 and 4 | 268 (109–457) | — | 140 (87–213) | — |
| E | Clade 4 | 194 (67–399) | — | 87 (42–147) | — |
| F | Clades 3a and 3b | 165 (63–292) | — | 92 (51–133) | — |
| G | Clade 2 | 161 (125–216) | 152 (131–177) | 160 (124–210) | 162 |
| H | Clade 1 | 140 (42–260) | — | 100 (49–164) | — |
| I | Clade 2b: all polar bears | 136 (120–164) | 134 (122–149) | 146 (120–179) | 136 |
| J | Clade 3a | 53 (21–95) | — | 49 (18–86) | — |
| K | Clade 3b | 42 (14–80) | — | 75 (43–113) | — |
| L | Clade 2b: extant polar bears | 41 (18–68) | 44 | — | 52 |
| M | Clade3a1 | 40 (15–72) | — | — | — |
| N | Clade 4: Southern Hokkaido | 36 (12–67) | — | — | — |
| O | Clade 2a | 31 (9–60) | 28 | 45 (10–91) | — |
| P | Clade 3a2 | 27 (10–49) | — | — | — |
Note.—Nodes refer to those in fig. 2. — indicates that no divergence time estimations are available. Node ages without parentheses have no information on 95% HPD.
aDivergence times were estimated based on the complete mtDNA sequences.
bDivergence times were estimated based on the short mtDNA control region sequences.
Discussion
Phylogeography
Our estimated TMRCA between the eastern and western lineages at approximately 566 kilo years before present (kyBP) (95% HPD: 251–944 kyBP) is consistent with the oldest fossil of the brown bear reported, from the early Pleistocene (Early Biharian, approximately 1.20 million years before present: MyBP) of Europe (Rabeder et al. 2010; Wagner and Čermák 2012). The three phylogenetically distinct maternal lineages of brown bears on Hokkaido are currently allopatrically distributed (fig. 1), as Matsuhashi et al. (1999) also found. This pattern suggests that three independent migrations to the island occurred. Hokkaido Island was connected with Sakhalin and the Eurasian Continent by land bridges until the end of the last glacial period at approximately 12 kyBP (Ohshima 1990). After the splits of the ancestral three lineages of Hokkaido brown bears on the Eurasian Continent, each lineage could have migrated to Hokkaido at different times before the disappearance of the land bridges. Tsugaru Strait between Hokkaido Island and Honshu Island of Japan was formed in the late Pleistocene approximately 100–150 kyBP (Ohshima 1990). Brown bears do not occur on Honshu Island at the present time, but brown bear fossils from mid- to late Pleistocene deposits have been reported from Honshu (Takakuwa et al. 2007). Our results (fig. 2) show that the ancestors of southern Hokkaido brown bears diverged from those of North American brown bears at approximately 194 kyBP (95% HPD: 67–399 kyBP) (fig. 2). After dispersal to Hokkaido bears from the southern Hokkaido population could then have dispersed southward to Honshu before Tsugaru Strait formed (approximately 100–150 kyBP); the Honshu population later went extinct.
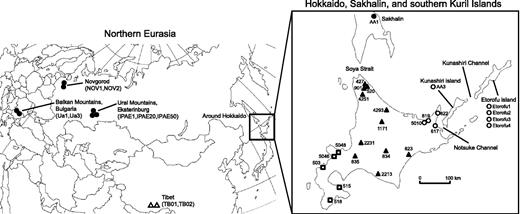
Geographical distribution of brown bear mtDNA clades on the Eurasian Continent and Hokkaido Island, Japan. Filled circles, mtDNA clade 3a1; filled triangles, clade 3a2; open circles, clade 3b; open squares, clade 4; open triangles, clade 5. Numbers next to the symbols are sample numbers.
Maximum clade credibility tree from a BEAST Bayesian analysis of complete mtDNA sequences from brown bears. The time scale was calibrated using both the radiocarbon dates of ancient bear sequences, which were previously reported (see the text). Major mtDNA clades and their geographic regions are labeled. Nodes of interest are those with a posterior probability of 1 or 0.99 or 0.98 in italics. The numbers at nodes A–P indicate mean ages in kyBP. Node bars represent the 95% highest posterior density of nodal age estimates. Terminal taxa are indicated by sample numbers or by accession numbers previously reported. Haplotype numbers in the parentheses for Hokkaido brown bear samples correspond to the mtDNA control region haplotypes of Matsuhashi et al. (1999). Slanted double lines near the root indicate that portions of lines or node bars have been omitted due to space constraints. Detailed information on nodal ages is given in table 1.
In this study, we analyzed complete mtDNA sequences, from which the matrilineal lineages of brown bears were inferred, and we discussed the migration history of the brown bear. If such complete mtDNA data sets are applied to population modeling (e.g., Approximate Bayesian Computation approaches), the analysis would reveal details of the migration history of the brown bear population. In our data set, however, the specimen sampling per geographical region was biased (i.e., brown bears from Hokkaido were broadly collected, whereas brown bears from the Eurasian continent were collected from limited areas). This bias may lead to the possibility that the migration history of the population is incorrectly estimated. Therefore, we did not carry out such an analysis, and only discussed molecular phylogenies and divergence time estimates. Further sampling of specimens from the Eurasian continent is needed.
Relationship between Brown Bears in Southern Hokkaido and Tibet
The southern Hokkaido brown bears grouped with North American brown bears (clade 4) rather than Tibetan brown bears (fig. 2; supplementary fig. S1, Supplementary Material online), a result strongly supported the relationship previously reported on Korsten et al. (2009), Davison et al. (2011), and Edwards et al. (2011). In contrast, it was inconsistent with that of Matsuhashi et al. (2001) based on 266-bp mtDNA control region sequences. These authors speculated that Tibetan brown bears might have been a relict population of the Eurasian brown bear, which in their results shared a common ancestor with the bears that colonized southern Hokkaido. The Tibetan brown bears were genetically distant from the rest of the eastern lineage, with an estimated divergence at approximately 343 kyBP (95% HPD: 139–582 kyBP) (fig. 2 and table 1; supplementary fig. S1, Supplementary Material online). This result suggests that Eurasian brown bears dispersed early into Tibet, and that the brown bears on the Tibetan Plateau subsequently remained geographically isolated from the source population.
The TMRCA between southern Hokkaido and North American brown bears (clade 4) at approximately 194 kyBP (95% HPD: 67–399 kyBP) indicates that the former could have moved onto Hokkaido first among the three Hokkaido lineages. Because the southern Hokkaido brown bears are closely related to the North American brown bears (clade 4) and distant from Tibetan brown bears, they possibly migrated onto Hokkaido via the land bridge connecting with Sakhalin and the Eurasian Continent. Other brown bears in clade 4 originating from the Eurasian Continent could have colonized North America via Beringia, because ancient brown bears at 36 kyBP in Alaska is confirmed to be classified into clade 4 (Leonard et al. 2000; Barnes et al. 2002).
In addition, this study shows that among southern Hokkaido brown bears, genetically more closely related mtDNA haplotypes are found in geographically closer locations (fig. 1; supplementary fig. S1, Supplementary Material online), compared with eastern and central Hokkaido brown bears, supporting the results of Matsuhashi et al. (1999) based on mtDNA control region data. This distribution pattern indicates that the phylogenetically more related haplotypes in clade 4 were appeared within southern Hokkaido and became fixed after immigration, and that the ancestor of the southern Hokkaido lineage was the first immigrated brown bear lineage on the island.
Relationships between Brown Bears in Eastern Hokkaido and the Southern Kuril Islands
After the divergence of clades 3a and 3b approximately 165 kyBP (95% HPD: 63–292 kyBP) (fig. 2 and table 1), representatives of clade 3b could have colonized Hokkaido via Sakhalin. Based on mtDNA cyt b and control region sequences, clade 3b includes brown bears from both eastern Hokkaido and eastern Alaska, suggesting that they descended from a MRCA on the Eurasian Continent (Matsuhashi et al. 2001; Korsten et al. 2009). The appearance of clade 3b brown bears at approximately 21 kyBP in the North American fossil record suggests that representatives of this clade colonized North America via Beringia (Barnes et al. 2002). Because no complete mtDNA sequences were available from eastern Alaskan brown bears in clade 3b; however, we did not estimate the overall TMRCA for clade 3b nor calculate the timing of the divergence between the eastern Hokkaido and eastern Alaskan lineages.
Individuals from Kunashiri and Etorofu Islands grouped with the eastern Hokkaido lineage (clade 3b) (fig. 2; supplementary fig. S1, Supplementary Material online), with the former populations more closely related to one another than either is to the Hokkaido population. This suggests that the Kunashiri and Etorofu populations originated through dispersal from eastern Hokkaido, a conclusion confirmed by the results of a craniometrical analysis (Baryshnikov and Puzachenko 2009). Kunashiri Island was connected to Hokkaido by a land bridge 8–110 kyBP, whereas Etorofu Island remained separate during that period (Igarashi 2000). Brown bears on Kunashiri likely became isolated from the eastern Hokkaido lineage when Notsuke Channel formed a sea barrier, and subsequently dispersed to Etorofu. Kunashiri Channel between Kunashiri and Etorofu is now approximately 25 km wide, though it could have been narrower during glacial maxima.
Relationships between Brown Bears in Central Hokkaido and on Sakhalin
In this study, clade 3a was subdivided into two geographically distinct clades: clade 3a1 comprising continentally widespread bears from eastern Europe, western Alaska, and Sakhalin; and clade 3a2 comprising local bears from central Hokkaido (fig. 2; supplementary fig. S1, Supplementary Material online). Clades 3a1 and 3a2 could have diverged on the Eurasian Continent, with a TMRCA of approximately 53 kyBP (95%HPD: 21–95 kyBP) for clade 3a. Clades 3a1 and 3a2 are differentiated by only five parsimony-informative nucleotide sites. The Kodiak brown bear (western Alaskan lineage) was more closely related to eastern European than to central Hokkaido bears, suggesting that this lineage dispersed to North America after divergence from the lineage leading to the central Hokkaido bears. Since the divergence time between clades 3a1 and 3a2 is approximately 53 kyBP (95% HPD: 21–95 kyBP), the ancestors of the central Hokkaido bears (clade 3a2) could have diverged earlier within clade 3a and dispersed into Hokkaido from the Eurasian Continent via Sakhalin before the appearance of Soya Strait between Hokkaido and Sakhalin/the Eurasian Continent approximately 12 kyBP (Ohshima 1990). On the other hand, western Alaskan brown bears grouped with eastern European brown bears, which means that representatives of clade 3a1 had migrated into North America after separation from the Eurasian population approximately 40 kyBP (95% HPD: 15–72 kyBP) but before formation of Bering Strait 11–13 kyBP (Elias et al. 1996). This is consistent with the lack of clade 3a brown bears among earlier Alaskan fossils (Leonard et al. 2000; Barnes et al. 2002). Two different colonization waves for clade 3a1 and 3a2 thus occurred at different times.
Although Sakhalin Island is considered to be a transient area for ancestors of the Hokkaido brown bears, the Sakhalin brown bear grouped with western Alaskan and eastern European brown bears in clade 3a1. This implies that the Sakhalin brown bear population did not originate at the same time as the dispersal of brown bears (clade 3a2) into central Hokkaido, but later during the geographical expansion of clade 3a1. Korsten et al. (2009) reported that brown bears in northern continental Eurasia (clade 3a) recently underwent a sudden expansion. Thus, representatives of clade 3a1 could have migrated onto Sakhalin separately from the other continental population on the island. One Sakhalin individual grouped with clade 3a1 in this study; however, there is a possibility that Sakhalin brown bears are composed of not only clade 3a1 brown bears but also other relict brown bears (i.e., clades 3a2, 3b, and 4) like Hokkaido brown bears. Further analyses of Sahkahin brown bears are needed and analyses of ancient DNA will also provide insights into the migration history of clades 3a1 and 3a2, but unfortunately no bear remains have yet been detected in Pleistocene strata on Sakhalin or Hokkaido.
Phylogenetic Relationships between Brown Bears and Polar Bears
Our study based on the maternally inherited mitochondrial genome showed the clade containing polar bears to be the sister group to the ABC islands brown bears; brown bears thus comprise a paraphyletic group. The TMRCAs for extant polar bears, all polar bears, and clade 2 (ABC islands brown bears and polar bears) were 41 kyBP (95% HPD: 18–68 kyBP), 136 kyBP (95% HPD: 120–164 kyBP), and 161 kyBP (95% HPD: 125–216 kyBP), respectively, indicating that the polar bear diverged from the ABC brown bear in the late Pleistocene. These results are remarkably similar to those of two studies based on the complete mtDNA genomes. Lindqvist et al. (2010) estimated corresponding TMRCAs at approximately 44, 134, and 152 kyBP, respectively, and Miller et al. (2012) also estimated corresponding TMRCAs at approximately 52, 136, and 162 kyBP, respectively. In contrast, in a study based on multiple, biparentally inherited nuclear loci, Hailer et al. (2012) estimated that polar bears diverged much earlier from brown bears, in the middle Pleistocene, approximately 600 kyBP, and that polar bears comprised the sister group to brown bears. Miller et al. (2012) highlighted the incongruence between the matrilineal and nuclear relationships among brown bears and polar bears. The discordant position of the ABC brown bears suggests an interspecific admixture as well as contrasting estimates for timing of divergence in the polar-brown bear lineage. The pronounced similarity in mtDNA characters between polar bears and brown bears may be explained by hybridization events in the Pleistocene (Edwards et al. 2011). To fully understand the evolutionary relationships between the two closely related species, it will be necessary to also analyze paternally inherited markers.
Materials and Methods
Samples and DNA Extraction
Muscle or liver samples from 20 brown bears collected on Hokkaido Island, Japan, were obtained from Environmental and Geological Research Department, Hokkaido Research Organization. Bear tissue samples were also obtained from the following regions and sources: four samples from Etorofu (Itrup) Island, one from Kunashiri (Kunashir) Island, one from southern Sakhalin, and two from Novgorod (Zoological Institute, Russian Academy of Sciences, St. Petersburg); three samples from the Ural Mountains (Institute of Plant and Animal Ecology, Russian Academy of Sciences, Ekaterinburg); two samples from the Balkan Mountains (Trakia University, Bulgaria); hairs from two Tibetan brown bears (Kobe Municipal Oji Zoo, Japan); and blood or muscle samples from four polar bears (Asahikawa Municipal Asahiyama Zoo, Japan). Sample numbers and geographical locations are shown in figure 1. Total genomic DNA was extracted using the DNeasy Tissue & Blood Kit (QIAGEN) or the QIAamp DNA Micro Kit (QIAGEN), following the manufacturer’s protocols.
DNA Amplification
Complete mtDNA sequences were amplified with a set of 11 primers reported in Delisle and Strobeck (2002) (supplementary table S1, Supplementary Material online). In the case of poor polymerase chain reaction (PCR) performance, a set of 13 newly designed primers was used (supplementary table S1, Supplementary Material online). PCR amplifications were performed in 20 μl reaction volumes each containing 2.0 µl of 10× PCR buffer (Takara), 1.6 μl of dNTP mixture (2.5 mM each dNTP; Takara), 0.1 μl of Taq DNA polymerase (5 U/μl; Takara), 0.1 μl each of forward and reverse primers (25 pmol/μl), 1.0 μl of DNA extract, and 14.9 μl of distilled water. Thermal cycling conditions were 3 min at 94 °C; 30–40 cycles of 1 min at 94 °C, 1 min at 50–60 °C, and 1.5 min at 72 °C; and a final extension for 10 min at 72 °C. For expected amplicon sizes more than 2 kb or for samples that showed poor PCR performance with Taq DNA polymerase (Takara), PCR amplifications were conducted in 20 μl reaction volumes containing 4.0 μl of 5× PrimeSTAR GXL DNA Buffer (Takara), 1.6 μl of dNTP mixture (2.5 mM each dNTP; Takara), 0.4 μl of PrimeSTAR GXL DNA Polymerase (1.25 U/μl, Takara), 0.2 μl each of forward and reverse primers (25 pmol/μl), 0.4 μl of bovine serum albumin (0.4 μg/μl), 1.0–4.0 μl of DNA extract, and 9.2–12.2 μl of distilled water. Thermal cycling conditions were 30–40 cycles of 10 s at 98 °C, 15 s at 50–60 °C, and 2 min at 68 °C. PCR products were purified with the QIAquick Purification Kit (Qiagen), following the manufacturer’s protocol.
Sequencing
DNA cycle sequencing was performed by using the BigDye v3.1 Cycle Sequencing Kit (Applied Biosystems, ABI) with the sequencing primers reported in Delisle and Strobeck (2002) or newly designed primers (supplementary tables S1 and Supplementary Data, Supplementary Material online). PCR for sequencing was performed in 10 μl volumes containing 1.75 μl of 5× BigDye Sequencing Buffer (ABI), 0.5 μl of Ready Reaction Premix (ABI), 1.0 μl of DNA template, and 5.15 μl of distilled water. Thirty cycles of (10 s at 96 °C, 5 s at 50 °C, and 4 min at 60 °C) were performed. Amplified DNA fragments were purified with isopropanol, and then formamide was added. Sequences were determined on an ABI 3730 DNA Analyzer. The exact length of the mtDNA control region was not determined due to the presence of variable-number tandem repeats (VNTRs). The genomic positions of two rRNAs, 22 tRNAs, 13 coding genes, and the control region were determined in relation to the complete mtDNA sequence of the brown bear (GenBank accession no. AF303110; Delisle and Strobeck 2002). DNA sequences generated in this study have been deposited in the DDBJ/NCBI/EMBL databases under accession nos. AP012559−012597.
Phylogenetic Analyses
Complete mtDNA sequences for nine brown bears (AF303110: Delisle and Strobeck 2002; EU497665: Bon et al. 2008; GU573486, GU573487, GU573489, and GU573491: Lindqvist et al. 2010; JX196367, JX196368, and JX196369: Miller et al. 2012), 27 extant polar bears (AF303111: Delisle and Strobeck 2002; AJ428577: Arnason et al. 2002; GU573485 and GU573490: Lindqvist et al. 2010; JX196370, JX196371, JX196372, JX196373, JX196374, JX196375, JX196376, JX196377, JX196378, JX196379, JX196380, JX196381, JX196382, JX196383, JX196384, JX196385, JX196386, JX196387, JX196388, JX196389, JX196390, JX196391, and JX196392: Miller et al. 2012), and one ancient polar bear (GU573488: Lindqvist et al. 2010) were obtained from GenBank and included in the alignment of sequences we obtained. The complete mtDNA sequence from one cave bear (Ursus spelaeus) (EU327344: Bon et al. 2008) was used as an outgroup. The sequence alignment was performed using Clustal W (Thompson et al. 1994) in MEGA 5.05 (Tamura et al. 2011). Insertions and deletions (indels), VNTRs consisting of 10-bp units in the control region, and ambiguous sites were excluded from analyses. NADH dehydrogenase subunit 6 (ND6) sequences were also excluded from the analyses because this gene is encoded on the strand (light strand) opposite in direction from the other 12 coding genes, and because its nucleotide composition is heterogeneous. The corrected alignment used for phylogenetic inference was 15,863-bp long and included two rRNA genes, 22 tRNA genes, 12 coding genes, and the control region (excluding VNTRs).
Phylogenetic trees were reconstructed by ML and BI. In both the ML and BI analyses, a partition model was used in which different substitution models were applied to different gene partitions. The best-fit substitution model was determined for each partition by using the Akaike information criterion for ML and the Bayesian information criterion (BIC) for BI, implemented in Kakusan 4 (Tanabe 2011). The ML tree was reconstructed using Treefinder version March 2011 (Jobb et al. 2004) and Phylogears 2 (Tanabe 2008) with 100 trials of the likelihood ratchet method (Vos 2003). Nodal support for the ML tree was assessed by bootstrap analysis with 1,000 pseudoreplicates. BI was performed with MrBayes v3.1.2 (Ronquist and Huelsenbeck 2003) in two simultaneous runs of 50,000,000 Markov chain Monte Carlo (MCMC) generations, with trees for estimation of the posterior probability distribution sampled every 100 generations; the first 5,000,000 trees were discarded as burn-in.
Estimation of Divergence Times
Divergence times were estimated with BEAST v1.6.2 (Drummond et al. 2007), with different substitution models applied to different gene partitions. The best-fit substitution model for each partition was estimated with the BIC implemented in Kakusan 4 (Tanabe 2011). The uncorrelated lognormal model was used to describe the relaxed clock. The constant size coalescent prior on the tree was used. Radiocarbon dates for ancient sequences were used to calibrate divergence ages for terminal nodes (internal calibration), including a mean age of 120 kyBP for the ancient polar bear (Lindqvist et al. 2010) and 31.87 kyBP for the cave bear (Bon et al. 2008). Posterior probability distributions of parameters including the trees were obtained by MCMC sampling. Trees were sampled every 10,000 generations from a total of 500,000,000 generations, with the first 50,000,000 generations discarded as burn-in. Acceptable mixing and convergence to the stationary distribution were checked by using Tracer v1.4 (Rambaut and Drummond 2007). The maximum clade credibility tree was produced with TreeAnnotator v1.6.2.
Acknowledgments
The authors thank Kobe Municipal Oji Zoo and Himeji City Zoo for providing specimens. They thank the staff of Asahikawa Municipal Asahiyama Zoological Park and Wildlife Conservation Center for their co-operation and support of sampling. They are also grateful to Prof. Matthew Dick for commenting on and editing the manuscript. They also thank M. A. Meshcheryakov, S. A. Piskunov, and S. I. Malygin for providing specimens. This work was supported in part by the Japan Society of the Promotion of Science for scientific research grant 17405012, Russian Foundation for Basic Research grant 13-04-00203, and Program “Biodiversity” of Russian Academy of Sciences.
References
Author notes
Associate editor: Emma Teeling