Abstract
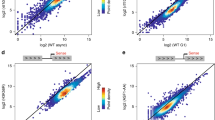
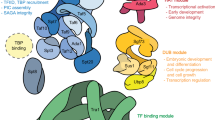
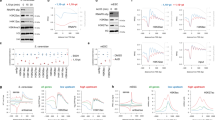
The transcription factors TFIID and SAGA are multi-subunit complexes involved in transcription by RNA polymerase II1,2. TFIID and SAGA contain common TATA-binding protein (TBP)-associated factor (TAFII) subunits and each complex contains a subunit with histone acetyltransferase activity3. These observations have raised questions about whether the functions of the two complexes in vivo are unique or overlapping. Here we use genome-wide expression analysis to investigate how expression of the yeast genome depends on both shared and unique subunits of these two complexes. We find that expression of most genes requires one or more of the common TAFII subunits, indicating that the functions of TFIID and SAGA are widely required for gene expression. Among the subunits shared by TFIID and SAGA are three histone-like TAFIIs, which have been proposed to form a sub-complex and mediate a common function in global transcription. Unexpectedly, we find that the histone-like TAFIIs have distinct roles in expression of the yeast genome. Most importantly, we show that the histone acetylase components of TFIID and SAGA (TAFII145 and Gcn5) are functionally redundant, indicating that expression of a large fraction of yeast genes can be regulated through the action of either complex.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hahn, S. The role of TAFs in RNA polymerase II transcription. Cell 95, 579–582 (1998).
Lee, T. & Young, R. Regulation of gene expression by TBP-associated proteins. Genes Dev. 12, 1398– 1408 (1998).
Grant, P. et al. A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation. Cell 94, 45–53 ( 1998).
Wodicka, L., Dong, H., Mittmann, M., Ho, M. H. & Lockhart, D. J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nature Biotechnol. 15, 1359 –1367 (1997).
Moqtaderi, Z., Bai, Y., Poon, D., Weil, P. A. & Struhl, K. TBP-associated factors are not generally required for transcriptional activation in yeast. Nature 383, 188–191 (1996).
Walker, S. S., Reese, J. C., Apone, L. M. & Green, M. R. Transcription activation in cells lacking TAFIIS. Nature 383, 185–188 (1996).
Walker, S. S., Shen, W. C., Reese, J. C., Apone, L. M. & Green, M. R. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell 90, 607–614 ( 1997).
Shen, W. & Green, M. Yeast TAF(II)145 functions as a core promoter selectivity factor, not a general coactivator. Cell 90, 615–624 (1997).
Apone, L. M., Virbasius, C. -M. A., Reese, J. C. & Green, M. R. Yeast TAF(II)90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 10 , 2368–2380 (1996).
Apone, L. et al. Broad, but not universal, transcriptional requirement for yTAFII17, a histone H3-like TAFII present in TFIID and SAGA. Mol. Cell 2, 653–661 (1998).
Holstege, F. C. P. et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95, 717–728 (1998).
Michel, B., Komarnitsky, P. & Buratowski, S. Histone-like TAFs are essential for transcription in vivo. Mol. Cell 2, 663– 673 (1998).
Moqtaderi, Z., Keaveney, M. & Struhl, K. The histone H3-like TAF is broadly required for transcription in yeast. Mol. Cell 2, 675– 682 (1998).
Natarajan, K., Jackson, B., Rhee, E. & Hinnebusch, A. yTAFII61 has a general role in RNA polymerase II transcription and is required by Gcn4p to recruit the SAGA coactivator complex. Mol. Cell 2, 683–692 (1998).
Sanders, S. L., Klebanow, E. R. & Weil, P. A. TAF25p, a non-histone-like subunit of TFIID and SAGA complexes, is essential for total mRNA gene transcription in vivo. J. Biol. Chem. 274, 18847–18850 (1999).
Komarnitsky, P. B., Michel, B. & Buratowski, S. TFIID-specific yeast TAF40 is essential for the majority of RNA polymerase II-mediated transcription in vivo. Genes Dev. 13, 2484–2489 ( 1999).
Website accompanying this paper. http://web.wi.mit.edu/young/TFIID_SAGA
Mizzen, C. A. et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87, 1261– 1270 (1996).
Hoffmann, A. et al. A histone octamer-like structure within TFIID. Nature 380, 356–359 ( 1996).
Xie, X. et al. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature 380, 316– 322 (1996).
Oelgeschlager, T., Chiang, C. M. & Roeder, R. G. Topology and reorganization of a human TFIID-promoter complex. Nature 382, 735– 738 (1996).
Horiuchi, J., Silverman, N., Pina, B., Marcus, G. & Guarente, L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol. Cell. Biol. 17, 3220–3228 (1997).
Roberts, S. M. & Winston, F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 3206–3213 ( 1996).
Roberts, S. M. & Winston, F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics 147, 451–465 ( 1997).
Sterner, D. et al. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19, 86–98 (1999).
Grant, P. et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11, 1640– 1650 (1997).
Eberharter, A. et al. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 19, 6621–6631 (1999).
Winston, F. & Sudarsanam, P. The SAGA of Spt proteins and transcriptional analysis in yeast: past, present, and future. Cold Spring Harb. Symp. Quant. Biol. 63, 553– 561 (1998).
Sudarsanam, P., Cao, Y., Wu, L., Laurent, B. & Winston, F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 18 , 3101–3106 (1999).
Biggar, S. & Crabtree, G. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 18 , 2254–2264 (1999).
Acknowledgements
We thank J. Workman for helpful discussions. This work was supported by funds from the NIH. F.C.P.H. was supported by a fellowship from the Human Frontier Science Program and E.G.J. is a predoctoral fellow of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Rights and permissions
About this article
Cite this article
Lee, T., Causton, H., Holstege, F. et al. Redundant roles for the TFIID and SAGA complexes in global transcription . Nature 405, 701–704 (2000). https://doi.org/10.1038/35015104
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/35015104
This article is cited by
-
Evaluation of reference genes for transcript analyses in Komagataella phaffii (Pichia pastoris)
Fungal Biology and Biotechnology (2023)
-
Human transcription factor protein interaction networks
Nature Communications (2022)
-
The Fungi-specific histone Acetyltransferase Rtt109 mediates morphogenesis, Aflatoxin synthesis and pathogenicity in Aspergillus flavus by acetylating H3K9
IMA Fungus (2021)
-
The SAGA of pineapple: genome-wide identification and tissue-specific expression of Spt-Ada-Gcn5-acetyltransferase (SAGA) complex
Euphytica (2021)
-
Disruption in phosphate transport affects membrane lipid and lipid droplet homeostasis in Saccharomyces cerevisiae
Journal of Bioenergetics and Biomembranes (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.