Abstract
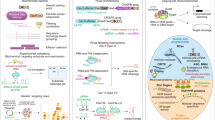
Now that the sequences of many genomes are available, methods are required for the rapid identification of functional genes. We describe here a simple system for the isolation of genes that function in the tumor necrosis factor-α (TNF-α)–mediated pathway of apoptosis, using RNA helicase–associated ribozyme libraries with randomized substrate-binding arms. Because target-site accessibility considerably limits the effective use of intracellular ribozymes, the effectiveness of a conventional ribozyme library has been low. To overcome this obstacle, we attached to ribozymes an RNA motif (poly(A)-tail) able to interact with endogenous RNA helicase(s) so that the resulting helicase-attached, hybrid ribozymes can more easily attack target sites regardless of their secondary or tertiary structures. When the phenotype of cells changes upon introduction of a ribozyme library, genes responsible for these changes may be identified by sequencing the active ribozyme clones. In the case of TNF-α-mediated apoptosis, when a ribozyme library was introduced into MCF-7 cells, surviving clones were completely or partially resistant to TNF-α-induced apoptosis. We identified many pro-apoptotic genes and partial sequences of previously uncharacterized genes using this method. Our gene discovery system should be generally applicable to the identification of functional genes in various systems.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
01 September 2006
A Correction to this paper has been published: https://doi.org/10.1038/nbt0906-1170e
References
Uhlenbeck, O.C. A small catalytic oligoribonucleotide. Nature 328, 596–600 (1987).
Haseloff, J. & Gerlach, W.L. Simple RNA enzymes with new and highly specific endonuclease activities. Nature 334, 585–591 (1988).
Symons, R.H. Small catalytic RNAs. Annu. Rev. Biochem. 61, 641–671 (1992).
Zhou, D.M. & Taira, K. The hydrolysis of RNA: from theoretical calculations to the hammerhead ribozyme–mediated cleavage of RNA. Chem. Rev. 98, 991–1026 (1998).
Krupp, G. & Gaur, R.K. Ribozyme: biochemistry and biotechnology. (Eaton Press, Natick; 2000).
Rossi, J.J. & Couture, L.A. Intracellular ribozyme applications. (Horizon Scientific Press, Norfolk; 1999).
Scanlon, K.J. Therapeutic applications of ribozymes. (Humana Press, Totowa; 1998).
Kawasaki, H. et al. Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature 393, 284–289 (1998).
Tanabe, T. et al. Oncogene inactivation in a mouse model: tissue invasion by leukaemic cells is stalled by loading them with a designer ribozyme. Nature 406, 473–474 (2000).
Lander, E.S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J.C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
van Berkum, N.L. & Holstege, F.C. DNA microarrays: raising the profile. Curr. Opin. Biotechnol. 12, 48–52 (2001).
Serebriiskii, I.G., Khazak, V. & Golemis, E.A. Redefinition of the yeast two-hybrid system in dialogue with changing priorities in biological research. Biotechniques 30, 634–636 (2001).
Li, Q.X., Robbins, J.M., Welch, P.J., Wong-Staal, F. & Barber, J.R. A novel functional genomics approach identifies mTERT as a suppressor of fibroblast transformation. Nucleic Acids Res. 28, 2605–2612 (2000).
Kruger, M. et al. Identification of eIF2Bγ and eIF2γ as cofactors of hepatitis C virus internal ribosome entry site-mediated translation using a functional genomics approach. Proc. Natl. Acad. Sci. USA 97, 8566–8571 (2000).
Beger, C. et al. Identification of Id4 as a regulator of BRCA1 expression by using a ribozyme-library-based inverse genomics approach. Proc. Natl. Acad. Sci. USA 98, 130–135 (2001).
Taira, K., Warashina, M., Kuwabara, T. & Kawasaki, H. Functional hybrid molecules with sliding ability. Japanese Patent Application H11-316133 (1999).
Beyaert, R. & Fiers, W. Molecular mechanisms of tumor necrosis factor–induced cytotoxicity. What we do understand and what we do not. FEBS Lett. 340, 9–16 (1994).
Heller, R.A. & Kronke, M. Tumor necrosis factor receptor–mediated signaling pathways. J. Cell Biol. 126, 5–9 (1994).
Lewis, M. et al. Cloning and expression of cDNAs for two distinct murine tumor necrosis factor receptors demonstrate one receptor is species-specific. Proc. Natl. Acad. Sci. USA 88, 2830–2834 (1991).
Schall, T.J. et al. Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell 61, 361–370 (1990).
Smith, C.A. et al. A receptor for tumor necrosis factor defines an unusual family of cellular and viral proteins. Science 248, 1019–1023 (1990).
Baker, S.J. & Reddy, E.P. Modulation of life and death by the TNF receptor superfamily. Oncogene 17, 3261–3270 (1998).
Hsu, H., Shu, H.B., Pan, M.G. & Goeddel, D.V. TRADD–TRAF2 and TRADD–FADD interactions define two distinct TNF receptor 1 signal transduction pathways. Cell 84, 299–308 (1996).
Boldin, M.P., Goncharov, T.M., Goltsev, Y.V. & Wallach, D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor–induced cell death. Cell 85, 803–815 (1996).
Cohen, G.M. Caspases: the executioners of apoptosis. Biochem. J. 326, 1–16 (1997).
Sun, X. M. et al. Distinct caspase cascades are initiated in receptor-mediated and chemical-induced apoptosis. J. Biol. Chem. 274, 5053–5060 (1999).
Kato, Y., Kuwabara, T., Warashina, M., Toda, H. & Taira, K. Relationships between the activities in vitro and in vivo of various kinds of ribozyme and their intracellular localization in mammalian cells. J. Biol. Chem. 276, 15378–15385 (2001).
Kuwabara, T. et al. A novel allosterically trans-activated ribozyme (maxizyme) with exceptional specificity in vitro and in vivo. Mol. Cell 2, 617–627 (1998).
Tang, H., Gaietta, G.M., Fischer, W.H., Ellisman, M.H. & Wong-Staal, F. A cellular cofactor for the constitutive transport element of type D retrovirus. Science 276, 1412–1415 (1997).
Craig, A.W., Haghighat, A., Yu, A.T. & Sonenberg, N. Interaction of polyadenylate-binding protein with the eIF4G homologue PAIP enhances translation. Nature 392, 520–523 (1998).
Warashina, M., Kuwabara, T., Kato, Y., Sano, M. & Taira, K. RNA–protein hybrid ribozymes that efficiently cleave any mRNA independently of the structure of the target RNA. Proc. Natl. Acad. Sci. USA 98, 5572–5577 (2001).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Whitehead, L., Kirk, H. & Kay, R. Expression cloning of oncogenes by retroviral transfer of cDNA libraries. Mol. Cell Biol. 15, 704–710 (1995).
Wittrup, K.D. Protein engineering by cell-surface display. Curr. Opin. Biotechnol. 12, 395–399 (2001).
Acknowledgements
This research was supported by grants from the Ministry of Economy, Trade and Industry (METI) of Japan, by a grant from the New Energy and Industrial Technology Development Organization (NEDO) of Japan, and by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
Kawasaki, H., Onuki, R., Suyama, E. et al. Identification of genes that function in the TNF-α-mediated apoptotic pathway using randomized hybrid ribozyme libraries. Nat Biotechnol 20, 376–380 (2002). https://doi.org/10.1038/nbt0402-376
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nbt0402-376
This article is cited by
-
Association of TNF-α-308 Polymorphism with Susceptibility to Autoimmune Hepatitis in Tunisians
Biochemical Genetics (2018)
-
TNF-α-induced Inflammation Stimulates Apolipoprotein-A4 via Activation of TNFR2 and NF-κB Signaling in Kidney Tubular Cells
Scientific Reports (2017)
-
Effects of 5-chloro-2-methyl-4-isothiazolin-3-one and other candidate biodiesel biocides on rat alveolar macrophages and NR8383 cells
Archives of Toxicology (2011)
-
Further accusations rock Japanese RNA laboratory
Nature (2006)
-
Retraction Note: Identification of genes that function in the TNF-α-mediated apoptotic pathway using randomized hybrid ribozyme libraries
Nature Biotechnology (2006)