Abstract
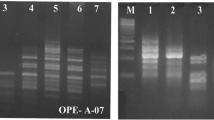
The genetic relationships among 10 inbred lines representing 10 populations of the autogamous annualMicroseris elegans from throughout California has been determined using random amplified polymorphic DNAs (RAPDs). Seventeen arbitrary 10 base pair primers produced 134 amplification products; 81 of these were shared by two or more strains. The 3 genotypes from Northern California are closely related as are 3 genotypes from Middle Californian populations which are not nearest neighbors. DNA fingerprinting with the oligonucleotide (GATA)4 gave compatible results, but the comparison was limited to samples run on one gel. Isoenzyme patterns are compatible with the DNA results, but limited by the very low number of informative polymorphisms. The clustered relationship among genotypes within a species and their geographic distribution suggests very restricted genetic recombination and an origin of new populations from randomly dispersed achenes within the range of the species.
Similar content being viewed by others
References
Ali, S., Müller, C. R., Epplen, J. T., 1986: DNA fingerprinting by oligonucleotide probes specific for simple repeats. — Hum. Genet.74: 239–243.
Bachmann, K., Chambers, K. L., 1990a: Heritable variation for heterocarpy inMicroseris bigelovii (Asteraceae—Lactuceae). — Beitr. Biol. Pflanzen65: 123–146.
—, —, 1990b: Genetic variation for the timing and site of trichomes on the leaves ofMicroseris bigelovii (Asteraceae: Lactuceae). — Biol. Zentralbl.109: 151–158.
—, —,Grau, J., Price, H. J., 1985a: Genetic variation inMicroseris pygmaea (Asteraceae—Lactuceae). — Beitr. Biol. Pflanzen60: 51–88.
—, —,Price, H. J., 1979: Genome size and phenotypic variation inMicroseris (Asteraceae, Cichorieae). — Pl. Syst. Evol. Suppl.2: 41–66.
—, —, —, 1984: Differential geographic distribution of spatulate and pointed leaf shapes inMicroseris bigelovii (Asteraceae—Lactuceae). — Beitr. Biol. Pflanzen59: 5–14.
—, —, —, 1982: Four additive genes determining pappus part numbers inMicroseris annual hybrid C34 (Asteraceae/Lactuceae). — Pl. Syst. Evol.141: 123–141.
—,Heusen, A. W. van, Chambers, K. L., Price, H. J., 1985b: Duplications of additively acting genes in the evolution of a plant (Microseris pygmaea). — Experientia41: 1348–1350.
—, —, —, —, 1987: Genetic variation for the onset of flowering inMicroseris bigelovii (Asteraceae, Lactuceae). — Beitr. Biol. Pflanzen62: 23–41.
Chambers, K. L., 1955: A biosystematic study of the annual spocies ofMicroseris. — Contrib. Dudley Herbarium4: 207–312.
Dellaporta, S. L., Wood, J., Hicks, J. B., 1983: A plant DNA minipreparation: version II. — Plant Mol. Biol. Reporter1: 19–21.
Epplen, J. T., 1988: On simple repeated GAt/cA sequences in animal genomes: a critical reappraisal. — J. Heredity79: 409–417.
Felsenstein, J., 1985: Confidence limits on phylogenies: an approach using the bootstrap. — Evolution39: 783–791.
Gebhardt, C., Blomendahl, C., Schachtschabel, U., Debener, T., Salamini, F., Ritter, E., 1989: Identification of 2n breeding lines and 4n varieties of potato (Solanum tuberosum, ssp.tuberosum) with RFLP fingerprints. — Theor. Appl. Genet.78: 16–22.
Gilbert, D. A., Lehman, N., O'Brien, S. J., Wayne, R. K., 1990: Genetic fingerprinting reflects population differentiation in the California Channel Island fox. — Nature344: 764–767.
Heusden, A. W. van, 1990: Genetic analysis of natural variation inMicroseris pygmaea (Asteraceae) in crosses withM. bigelovii C93b. — Thesis, Amsterdam.
—,Rouppe van der Voort, J., Bachmann, K., 1991: Oligo-(GATA) fingerprints identify clones in asexual dandelions (Taraxacum, Asteraceae). — Fingerprint News3(2: 13–15.
Houten, W. H. J. van, Heusden, A. W. van, Rouppe van der Voort, J., Raijmann, L., Bachmann, K., 1991: Hypervariable DNA fingerprint loci inMicroseris pygmaea (Asteraceae, Lactuceae). — Bot. Acta104: 252–256.
Jeffreys, A. J., Wilson, V., Thein, S. L., 1985: Individual specific fingerprints of human DNA. — Nature316: 76–79.
Maniatis, T., Fritsch, E. F., Sandbrook, J., 1982: Molecular cloning: a laboratory manual. — New York: Cold Spring Harbor Laboratory.
Nybom, H., Rogstad, S. H., 1990: DNA “fingerprints” detect genetic variation inAcer negundo (Aceraceae). — Pl. Syst. Evol.173: 49–56.
—, —,Schaal, B. A., 1990: Genetic variation detected by use of the M13 “DNA fingerprint” probe inMalus, Prunus, andRubus (Rosaceae). — Theor. Appl. Genet.79: 153–156.
—,Schaal, B. A., Rogstad, S. H., 1989: DNA “fingerprints” can distinguish cultivars of blackberries and raspberries. — Acta Hortic.262: 310–315.
Sneath, P. H. A., Sokal, R. R., 1973: Numerical taxonomy. — San Francisco: Freeman.
Wallace, R. S., Jansen, R. K., 1990: Systematic implications of chloroplast DNA variation in the genusMicroseris (Asteraceae: Lactuceae). — Syst. Bot.15: 606–616.
Weising, K., Kahl, G., 1990: DNA fingerprinting in plants — the potential of a new method. — Biotech-Forum Europe7: 230–235.
Williams, J. K. G., Kubelik, A. R., Litvak, K. J., Rafalski, J. A., Tingey, S. V., 1990: DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. — Nucleic Acids Res.18: 6531–6535.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
van Heusden, A.W., Bachmann, K. Genotype relationships inMicroseris elegans (Asteraceae, Lactuceae) revealed by DNA amplification from arbitrary primers (RAPDs). Pl Syst Evol 179, 221–233 (1992). https://doi.org/10.1007/BF00937598
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00937598