Abstract
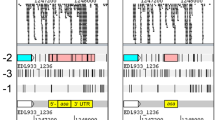
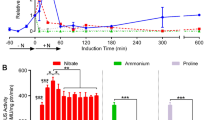
Three overlapping clones covering a Chlarnydomonas reinhardtii genomic region of about 32 kb appear to contain five genes potentially involved in nitrate assimilation in addition to the nitrate reductase structural locus nit-1. These new loci produced transcripts of 2.8, 2.2, 1.8 and 1.7 kb in nitrate-induced wild-type cells that, like the 3.4 kb transcript of nit-1, were undetectable in cells grown in ammonium. In addition, in a mutant defective at the regulatory locus, nit-2 for nitrate assimilation, which does not express the nit-1 gene transcript, accumulation of the four other transcripts was also blocked. They have been named nar (nitrate assimilation related) genes. The nar-1 and nar-2 loci are transcribed in the same orientation as nit-1. The nar-3 and nar-4 loci are transcribed divergently from nit-1. DNA and RNA sequences from both nar-3 and nar-4 cross-hybridized with each other indicating that they share similar sequences. Four nitrate assimilation-deficient mutants (C2, D2, F6 and G1) were characterized. These mutants lack nar transcripts and have major deletions and/or rearrangements in the nar gene cluster. In contrast to other nitrate reductase-deficient mutants and to wild type, deletion mutants and the regulatory mutant nit-2 were incapable of accumulating intracellular nitrate. Two of the mutants in which expression of all of the nar loci did not occur, C2 and D2, grew in nitrite medium and showed wild-type levels of both nitrite uptake and nitrite reductase activities. Thus the nar loci cannot be required for nitrite assimilation. Mutants F6 and G1 were unable to grow in either nitrite- or nitrate-containing medium, and lacked nitrate reductase, nitrite reductase, nitrate uptake and nitrite uptake activities. The inability to assimilate nitrite co-segregated with nit-1 in crosses between these mutants and wild type. These results indicate that a complex gene cluster responsible for the assimilation of nitrate has been identified in C. reinhardtii, and that, in addition, at least one locus necessary for nitrite assimilation is genetically linked to this cluster.
Similar content being viewed by others
References
Arnon DI (1949) Copper enzymes in isolated chloroplast. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Arrand JE (1985) Preparation of nucleic acid probes. In: Hames BD, Higgins SJ (eds) Nucleic Acid Hybridization: A Practical Approach. IRL Press, Oxford, pp 17–45
Back E, Burkhart W, Moyer M, Privalle L, Rothstein S (1988) Isolation of cDNA clones coding for spinach nitrite reductase: complete sequence and nitrate induction. Mol Gen Genet 212:20–26
Benton WD, Davis RW (1977) Screening λgt recombinant clones by hybridization to single plaques in situ. Science 196:180–182
Caboche M, Rouzé P (1990) Nitrate reductase: a target for molecular and cellular studies in higher plants. Trends Genet 6:187–192
Córdoba F, Cárdenas J, Fernández E (1986) Kinetic characterization of nitrite uptake and reduction by Chlamydomonas reinhardtii. Plant Physiol 82:904–908
Crawford NM, Campbell WH (1990) Fertile fields. Plant Cell 2:829–835
Feinberg AP, Vogelstein B (1984) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 137:266–267
Fernández E, Cárdenas J (1981) Occurrence of xanthine dehydrogenase in Chlamydomonas reinhardtii: A common cofactor shared by xanthine dehydrogenase and nitrate reductase. Planta 153:254–257
Fernández E, Cárdenas J (1989) Genetics and regulatory aspects of nitrate assimilation in algae. In: Wray JL, Kinghorn JR (eds) Molecular and Genetic Aspects of Nitrate Assimilation. Oxford University Press, Oxford, pp 101–124
Fernández E, Matagne RF (1984) Genetic analysis of nitrate reductase-deficient mutants in Chlamydomonas reinhardtii. Curr Genet 8:635–640
Fernández E, Schnell R, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA (1989) Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 86:6449–6453
Florencio FJ, Vega JM (1983) Utilization of nitrate, nitrite and ammonium by Chlamydomonas reinhardtii: Photoproduction of ammonium. Planta 158:288–293
Galván A, Cárdenas J, Fernández E (1992) Nitrate reductase regulates expression of nitrite uptake and nitrite reductase activities in Chlamydomonas reinhardtii. Plant Physiol 98:422–426
Guerrero MG, Vega JM, Losada M (1981) The assimilatory nitratereducing system and its regulation. Annu Rev Plant Physiol 32:169–204
Harris EH (1989) The Chlamydomonas Sourcebook. Academic Press, New York
Johnstone IL, McCabe PC, Greaves P, Gurr SJ, Cole GE, Brow MAD, Unkles SE, Clutterbuck AJ, Kinghorn JR, Innis MA (1990) Isolation and characterization of the crnA-niiA-niaD gene cluster for nitrate assimilation in Aspergillus nidulans. Gene 90:181–192
Kunlemeier C, Green PJ, Chua NH (1987) Regulation of gene expression in higher plants. Annu Rev Plant Physiol 38:221–257
Levine RP, Ebersold WT (1960) The genetics and cytology of Chlamydomonas. Annu Rev Microbiol 14:197–216
McCabe AP, Riach MBR, Unkles SE, Kinghorn JR (1990) The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J 9:279–287
McClure BA, Hagen G, Brown CS, Gee MA, Guilfoyle TJ (1989) Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1:229–239
Pineda M, Fernández E, Cárdenas J (1984) Urate oxidase of Chlamydomonas reinhardtii. Physiol Plant 62:453–457
Ranum LPW, Thompson MD, Schloss JA, Lefebvre PA, Silflow CD (1988) Mapping flagellar genes in Chlamydomonas using restriction fragment length polymorphisms. Genetics 120:109–122
Romero LC, Galván F, Vega JM (1987) Purification and properties of the siroheme-containing ferredoxin nitrite reductase from Chlamydomonas reinhardtii. Biochim Biophys Acta 914:55–63
Romero JM, Lara C, Guerrero MG (1989) Determination of intracellular nitrate. Biochem J 259:545–548
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: A laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor New York
Schloss JA, Silflow CD, Rosenbaum JL (1984) mRNA abundance changes during flagellar regeneration in Chlamydomonas reinhardtii. Mol Cell Biol 4:424–434
Snell FD, Snell CT (1949) Colorimetric methods of analysis, vol 2. Van Nostrand, New York, pp 804–907
Sutliff TD, Huang N, Litts JC, Rodriguez RL (1991) Characterization of an α-amylase multigene cluster in rice. Plant Mol Biol 16:579–591
Unkles SE (1988) Fungal biotechnology and the nitrate assimilation pathway. In: Wray JL, Kinghorn JR (eds) Molecular and genetic aspects of nitrate assimilation. Oxford University Press, Oxford, pp 341–363
Unkles SE, Hawker KL, Campbell EI, Montague P, Kinghorn JR (1991) crnA encodes a nitrate transporter in Aspergillus nidulans. Proc Natl Acad Sci USA 88:204–208
Van Winkle-Swift KP (1977) Maturation of algal zygots: alternative experimental approaches for Chlamydomonas reinhardtii. J Phycol 13:225–231
Vega JM, Herrera J, Aparicio PJ, Paneque A, Losada M (1971) Role of molybdenum in nitrate reduction by Chlorella. Plant Physiol 48:292–299
Wray JL, Kinghorn JR (1989) Molecular and genetic aspects of nitrate assimilation. Oxford University Press, Oxford
Youngblom J, Schloss JA, Silflow CD (1984) The two β-tubulin genes of Chlamydomonas reinhardtii code for identical proteins. Mol Cell Biol 4:2686–2696
Zimmer WE, Schloss JA, Silflow CD, Youngblom J, Watterson DM (1988) Structural organization, DNA sequence, and expression of the calmodulin gene. J Biol Chem 263:19370–19383
Author information
Authors and Affiliations
Additional information
Communicated by R.G. Herrmannández
Rights and permissions
About this article
Cite this article
Quesada, A., Galván, A., Schnell, R.A. et al. Five nitrate assimilation-related loci are clustered in Chlamydomonas reinhardtii . Molec. Gen. Genet. 240, 387–394 (1993). https://doi.org/10.1007/BF00280390
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00280390