Abstract
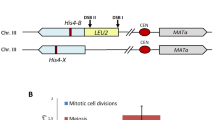
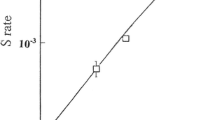
We have employed the analysis of spontaneous forward mutations that confer the ability to utilize L-α-aminoadipate as a nitrogen source (α-Aa+) to discern the events that contribute to mitotic segregation of spontaneous recessive mutations by diploid cells. α-Aa- diploid cells yield α-Aa+ mutants at a rate of 7.8±3.6×10-9. As in haploid strains, approximately 97% (30/31) of α-Aa+ mutants are spontaneous lys2-x recessive mutations. α-Aa+ mutants of diploid cells reflect mostly the fate of LYS2/lys2-x heterozygotes that arise by mutation within LYS2/LYS2 populations at a rate of 1.2±0.4×10-6. Mitotic recombination occurs in nonrandom association with forward mutation of LYS2 at a rate of 1.3±0.6×10-3. This mitotic recombination rate is tenfold higher than that of a control LYS2/lys2-1 diploid. Mitotic segregation within LYS2/lys2-x subpopulations yields primarily lys2-x/lys2-x diploids and a minority of lys2-x aneuploids. Fifteen percent of lys2-x/lys2-x diploids appear to have arisen by gene conversion of LYS2 to lys2-x; 85% of lys2-x/lys2-x diploids appear to have arisen by mitotic recombination in the CENII-LYS2 interval. lys2-1/lys2-1 mitotic segregants of a control LYS2/lys2-1 diploid consist similarly of 18% of lys2-1/lys2-1 diploids that appear to have arisen by gene conversion of LYS2 to lys2-1 and 82% of lys2-1/lys2-1 diploids that appear to have arisen by mitotic recombination in the CENII-LYS2 interval. The methods described can be used to simultaneously monitor the effects of yeast gene mutations and carcinogens on the principal parameters affecting the genomic stability of diploid mitotic cells: mutation, gene conversion, intergenic recombination, and chromosomal loss or rearrangement.
Similar content being viewed by others
References
Chatoo BB, Sherman F, Azubalis DA, Fjellstadt TA, Mehnert D, Ogur M (1979) Genetics 93:51–56
Duin M van, de Wit J, Odijk H, Westerveld A, Yasui A, Koken MHM, Hoeijmakers JHJ, Bootsma D (1986) Cell 44:913–923
Esposito MS, Brown JT (1990) Curr Genet 17:7–12
Esposito MS, Bruschi CV (1993) Curr Genet 23:430–434
Esposito MS, Maleas D, Bjornstad K, Holbrook LL (1986) Curr Genet 10:425–433
Esposito MS, Ramirez RM, Bruschi CV (1994) Curr Genet 25: 1–11
Friedberg EC, Siede W, Cooper AJ (1991) In: Broach JR, Pringle JR, Jones EW (eds) The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics vol 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp 147–192
Golin JE, Esposito MS (1981) Mol Gen Genet 183:252–263
Haynes RH, Kunz BA (1981) In: Strathern JN, Jones EW, Broach J (eds) The molecular biology of the yeast Saccharomyces: life cycle and inheritance. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp 371–414
Heude M, Fabre F (1993) Genetics 133:489–498
Klein F, Karwan A, Wintersberger U (1989) Mutat Res 210:157–164
Klein F, Karwan A, Wintersberger U (1990) Genetics 124:57–65
Lea DE, Coulson CA (1949) J Genet 49:264–285
Luria S, Delbruck M (1943) Genetics 28:491–511
Mortimer RK, Schild D (1985) Microbiol Rev 49:181–212
Palombo F, Hughes M, Jiricny J, Truong O, Hsuan J (1994) Nature 367:417
Parry JM, Zimmermann FK (1976) Mutat Res 36:49–66
Petes TD, Malone RE, Symington LS (1991) In: Broach JR, Pringle JR, Jones EW (eds) The molecular and cellular biology of the yeast Saccharomyces: genome dynamics, protein synthesis and energetics vol 1. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York, pp 407–521
Roman H (1956) Cold Spring Harbor Symp Quant Biol 21:175–185
Roman H, Ruzinski MM (1990) Genetics 124:7–25
Sager R (1989) Science 246:1406–1412
Solomon E, Borrow J, Goddard AD (1991) Science 254:1153–1160
Thomson G (1988) Annu Rev Genet 22:31–50
Author information
Authors and Affiliations
Additional information
Communicated by P. P. Slonimski
The research of the authors was supported by the Director, Office of Energy Research, Biological Research Division of the U.S. Department of Energy under Contract No. DE-ACO3-76SF00098, grants to M. S. E. and C. V. B. from the National Institutes of Health and the National Aeronautics and Space Administration, and a National Science Foundation postdoctoral research fellowship award to R. M. R.
Rights and permissions
About this article
Cite this article
Esposito, M.S., Ramirez, R.M. & Bruschi, C.V. Nonrandomly-associated forward mutation and mitotic recombination yield yeast diploids homozygous for recessive mutations. Curr Genet 26, 302–307 (1994). https://doi.org/10.1007/BF00310493
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00310493