Abstract
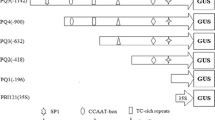
The 5′- and 3′-flanking regions of HPRA, a cucumber gene that encodes hydroxypyruvate reductase, were evaluated for regulatory activity with respect to light responsiveness and organ specificity. To define the functional regions of the 5′-flanking region of HPRA, a series of deletions was generated and the remaining portions fused to the β-glucuronidase (GUS) reporter gene (uidA) containing a minimal 35S promoter truncated at −90. The region from −66 to +39 was found to be necessary for light-regulated expression of the uidA reporter gene, while the region from −382 to −67 was found to be necessary for its leaf-specific expression. Further deletion of the HPRA 5′ flanking region to −590 resulted in high levels of root expression, suggesting the presence of a negative regulatory element responsible for silencing root expression of the HPRA gene between −590 and −383.
The 3′-flanking region of the HPRA gene downstream of the polyadenylation site contains several sequence motifs resembling regulatory elements present in the promoters of several light-responsive genes. An 823 bp portion of the HPRA 3′-flanking region containing these putative regulatory elements enhanced GUS expression in leaves when placed downstream of the uidA reporter gene in the forward orientation, but not in the reverse orientation. When placed 5′ of the −90 35S promoter, the 823 bp fragment enhanced slightly, independently of orientation, the root tip-specific expression pattern intrinsic to the −90 35S promoter, indicating that in some cases this region can act as a transcriptional enhancer.
Similar content being viewed by others
References
An G, Ebert PR, Mitra A, Ha SB: Binary vectors. In: Gelvin SB, Schilperoort RA, Verma DPS (eds) Plant Molecular Biology Manual, pp. A3/1-A3/16. Kluwer Academic Publishers, Dordrecht (1988).
Benfey PN, Ren L, Chua N-H: Organ-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J 9: 1677–1684 (1990).
Benfey PN, Ren L, Chua N-H: Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J 9: 1685–1696 (1990).
Bertoni G, Becker WM: Effects of light fluence and wavelength on expression of the gene encoding cucumber hydroxypyruvate reductase. Plant Physiol 103: 933–941 (1993).
Bevan M: Binary Agrobacterium vectors for plant transformation. Nucl Acids Res 12: 8711–8721 (1984).
Bodine DM, Ley TJ: An enhancer element lies 3′ to the human Ag globin gene. EMBO J 6: 2997–3004 (1987).
Bradford MM: A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 (1976).
Choi O-R, Engel JD: A 3′ enhancer is required for temporal and organ-specific transcriptional activation of the chicken adult β-globin gene. Nature 323: 731–734 (1986).
Datta N, Cashmore AR: Binding of a pea nuclear protein to promoters of certain photoregulated genes is modulated by phosphorylation. Plant Cell 1: 1069–1077 (1989).
Dean C, Favreau M, Bedbrook J, Dunsmuir P: Sequences 5′ to translation start regulate expression of petunia rbcS genes. Plant Cell 1: 209–215 (1989).
Dean C, Favreau M, Bond-Nutter D, Bedbrook J, Dunsmuir P: Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell 1: 201–208 (1989).
Dickey LF, Gallo-Meagher M, Allen GC, Thompson WF: Light regulatory sequences are located within the 5′ portion of the Fed-1 message sequence. EMBO J 11: 2311–2317 (1992).
Dietrich RA, Radke SE, Harada JJ: Downstream sequences are required to active a gene expressed in the root cortex of embryos and seedlings. Plant Cell 4: 1371–1382 (1992).
Douglass CJ, Hauffe KD, Ites-Morales ME, Ellard M, Paszkowski U, Hahlbrock K, Dangl JL: Exonic sequences are required for elicitor and light activation of a plant defense gene, but promoter sequences are sufficient for tissue specific expression. EMBO J 10: 1767–1775 (1991).
Doyle JJ, Doyle JL: Isolation of plant DNA from fresh tissue. Focus 12: 13–15 (1990).
Elliott RC, Dickey LF, White MJ, Thompson WF: Cis-acting elements for light regulation of pea ferredoxin I gene expression are located within transcribed sequences. Plant Cell 1: 691–698 (1989).
Feinberg AP, Vogelstein B: A technique for radiolabeling restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 (1983).
Fluhr R, Kuhlemeier C, Nagy F, Chua N-H: Organ-specific and light-induced expression of plant genes. Science 232: 1106–1112 (1986).
Gallagher SR: GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, pp. 47–59. Academic Press, San Diego (1992).
Gallo-Meagher M, Sowinski D, Elliott RC, Thompson WF: Both internal and external regulatory elements control expression of the pea Fed-1 gene in transgenic tobacco seedlings. Plant Cell 4: 389–395 (1992).
Gilmartin PM, Chua N-H: Spacing between GT-1 binding sites within a light-responsive element is critical for transcriptional activity. Plant Cell 2: 447–455 (1990).
Gilmartin PM, Sarokin L, Memelink J, Chua N-H: Molecular light switches for plant genes. Plant Cell 2: 369–378 (1990).
Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR: An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093 (1988).
Green PJ, Kay SA, Chua N-H: Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6: 2543–2549 (1987).
Green PJ, Yong M-H, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua N-H: Binding site requirements for pea nuclear factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A. EMBO J 7: 4035–4044 (1988).
Greenler JMcC, Sloan JS, Schwartz BW, Becker WM: Isolation, characterization and sequence analysis of a full-length cDNA clone encoding NADH-dependent hydroxypyruvate reductase from cucumber. Plant Mol Biol 13: 139–150 (1989).
Greenler JMcC, Becker WM: Organ specificity and light regulation of NADH-dependent hydroxypyruvate reductase transcript abundance. Plant Physiol 94: 1484–1487 (1990).
Henikoff S: Unidirectional digestion with exonuclease III in DNA sequence analysis. Meth Enzymol 155: 156–165 (1987).
Hoekema A, Hirsch PR, Hooykaas PJJ, Schilperoort RA: A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303: 179–180 (1983).
Hondred D, Wadle DM, Titus DE, Becker WM: Light-stimulated accumulation of the peroxisomal enzymes hydroxypyruvate reductase and serine: glyoxylate aminotransferase and their translatable mRNAs in cotyledons of cucumber seedlings. Plant Mol Biol 9: 259–275 (1987).
Hooykaas PJJ: Agrobacterium molecular genetics. In: Gelvin SB, Schilperoort RA (eds) Plant Molecular Biology Manual, A4, pp. 1–13. Kluwer Academic Publishers, Dordrecht (1988).
Husic DW, Husic HD, Tolbert NE: The oxidative photosynthetic carbon cycle or C2 cycle. CRC Crit Rev Plant Sci 5: 45–100 (1987).
Jarrell KA, Meselson M: Drosophila retrotransposon promoter includes an essential sequence at the initiation site and requires a downstream sequence for full activity. Proc Natl Acad Sci USA 88: 102–104 (1991).
Kagawa T, Beevers H: The development of microbodies (glyoxysomes and leaf peroxisomes) in cotyledons of germinating wheat seedlings. Plant Physiol 55: 258–264 (1975).
Kay SA, Keith B, Shinozaki K, Chye M-L, Chua N-H: The rice phytochrome gene: Structure, autoregulated expression, and binding of GT-1 to a conserved site in the 5′ upstream region. Plant Cell 1: 351–360 (1989).
Kuhlemeier C, Cuozzo M, Green PJ, Goyvaerts E, Ward K, Chua N-H: Localization and conditional redundancy of regulatory elements in rbcS-3A, a pea gene encoding the small subunit of ribulose-bisphosphate carboxylase. Proc Natl Acad Sci USA 85: 4665–4666 (1988).
Lam E, Benfey PN, Gilmartin PM, Fang R-X, Chua N-H: Site-specific mutations alter the in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci USA 86: 7890–7894 (1989).
Maniatis T, Fritsch EF, Schell J: Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Press, Cold Spring Harbor, NY (1982).
Martin T, Wohner R-V, Hummel S, Willmitzer L, Frommer WB: The GUS reporter system as a tool to study plant gene expression. In: Gallagher SR (ed) GUS Protocols: Using the GUS Gene as a Reporter of Gene Expression, pp. 26–27. Academic Press, San Diego (1992).
Nap J-P, Keizer P, Jansen R: First-generation transgenic plants and statistics. Plant Mol Biol Rep 11: 156–164 (1993).
Rogers SG, Horsch RB, Fraley RT: Gene transfer in plants: Production of transformed plants using Ti plasmid vectors. Meth Enzymol 118: 627–640 (1986).
Sanger F, Coulson AR, Barrell BG, Smith AJH, Roe BA: Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol 143: 161–178 (1980).
Schindler U, Cashmore AR: Photoregulated gene expression may involve ubiquitous DNA binding proteins. EMBO J 9: 3415–3427 (1990).
Schnarrenberger C, Oeser A, Tolbert NE: Development of microbodies in sunflower cotyledons and castor bean endosperm during germination. Plant Physiol 48: 566–574 (1971).
Schöpfer P, Apel K: Intracellular photomorphogenesis. In: Schropshire W, Mohr H (eds) Encyclopedia of Plant Physiology, vol. 16A, pp. 258–288. Springer-Verlag, Berlin (1983).
Schwartz BM, Sloan JS, Becker WM: Characterization of genes encoding hydroxypyruvate reductase in cucumber. Plant Mol Biol 17: 941–947 (1991).
Schwartz BW: Cloning and analysis of regulation of a cucumber gene encoding NADH-dependent hydroxypyruvate reductase. Ph.D. thesis, Department of Genetics, University of Wisconsin-Madison (1992).
Sloan JS, Schwartz BW, Becker WM: Promoter analysis of a light-regulated gene encoding hydroxypyruvate reductase, an enzyme of the photorespiratory glycolate pathway. Plant J 3: 867–874 (1993).
Stafford HA, Magaldi A, Vennesland B: The enzymatic reduction of hydroxypyruvic acid to D-glyceric acid in higher plants. J Biol Chem 207: 621–629 (1954).
Stockhaus J, Schell J, Willmitzer L: Correlation of the expression of the nuclear photosynthetic gene ST-LS1 with the presence of chloroplasts. EMBO J 8: 2445–2451 (1989).
Trainor CD, Stamler SJ, Engel DJ: Erythroid-specific transcription of the chicken histone H5 gene is directed by a 3′ enhancer. Nature 328: 827–830 (1987).
Trelease RN, Becker WM, Gruber PJ, Newcomb EH: Microbodies (glyoxysomes and peroxisomes) in cucumber cotyledons. Plant Physiol 48: 461–475 (1971).
Ueda T, Pichersky E, Malik VS, Cashmore AR: Level of expression of the tomato rbcS-3A gene is mediated by a far upstream promoter element in a developmentally regulated manner. Plant Cell 1: 217–227 (1989).
Vieria J, Messing J: The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19: 259–268 (1982).
Yanisch-Perron C, Vieria J, Messing J Improved: M13 phage cloning vectors and host strains: Nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33: 103–119 (1985).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Daniel, S.G., Becker, W.M. Transgenic analysis of the 5′- and 3′-flanking regions of the NADH-dependent hydroxypyruvate reductase gene from Cucumis sativus L.. Plant Mol Biol 28, 821–836 (1995). https://doi.org/10.1007/BF00042068
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00042068