Metabarcoding and microscopy characterization of phytoplankton from frontal areas of the Argentine Sea
- 1División Ficología, Facultad de Ciencias Naturales y Museo, Universidad Nacional de La Plata, La Plata, Argentina
- 2Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Ciudad Autónoma de Buenos Aires, Argentina
- 3Polar Biological Oceanography, Alfred Wegener Institut-Helmholtz Zentrum für Polar- und Meeresforschung, Bremerhaven, Germany
- 4Centro Austral de Investigaciones Científicas (CADIC), CONICET, Ushuaia, Argentina
- 5Fundación Bariloche, San Carlos de Bariloche, Argentina
- 6ZeroCarbono, Santiago, Chile
The Argentine Sea is worldwide recognized as a highly productive area, characterized by extensive phytoplankton blooms during spring and summer. Despite the well-known importance of frontal areas for biomass accumulation, phytoplankton diversity remains poorly studied. In an unprecedented approach for the Argentine Sea, we combined microscopy and 18Sv4 metabarcoding analyses for a refined assessment of summer phytoplankton composition in three understudied frontal areas of the Argentine Sea (≈43°−55°S), with contrasting oceanographic conditions. Metabarcoding and microscopy analyses agreed on the detection of the dominant phytoplanktonic groups in the different frontal areas studied; chlorophytes in Valdés Peninsula, dinoflagellates in waters off Blanco Cape, and diatoms in de los Estados Island. The analysis of the phytoplankton community was significantly enriched by combining both techniques, microscopy provided cell abundances and biomass data and metabarcoding provided greater detail on species composition, revealing an important specific richness of dinoflagellates, diatoms and other delicate groups, such as chlorophytes. However, we also considered differences between the methods for certain taxa at a lower taxonomic level (species/genus) of the dominant taxa, such as the underestimation of the diatoms Asterionellopsis glacialis and Pseudo-nitzschia spp. and the overestimation of Chaetoceros contortus by metabarcoding in comparison to microscopic counts. The detection of several taxa belonging to small and delicate groups, previously overlooked due to the lack of distinct morphological features, establishes a baseline for future studies on phytoplankton diversity in the Argentine Sea.
1 Introduction
The Argentine Sea, located in the southwestern Atlantic Ocean (≈37–55°S), is recognized as an area of high phytoplankton biomass accumulation based on multi-year satellite observations (Rivas et al., 2006). Large phytoplankton blooms occur during austral spring and summer, mainly associated with the occurrence of oceanographic fronts (Acha et al., 2004; Piola, 2008). High chlorophyll a (Chl-a) concentrations have been mostly associated with diatoms and dinoflagellates (Carreto et al., 2016; Gonçalves-Araujo et al., 2016; Guinder et al., 2018; Nunes et al., 2019), and also with coccolithophorids (García et al., 2011). Blooms of small (2–5 μm) flagellated or coccoid cells has also been observed, during late summer (Almandoz et al., 2007; Silva et al., 2009; Antacli et al., 2018), although their taxonomic identity has not been clearly resolved.
Several microscopy-based studies revealed a high diatom and dinoflagellate diversity in different regions of the Argentine sea (Balech, 1988; Cefarelli et al., 2010; Antacli et al., 2018). Moreover, detailed taxonomic studies, based on molecular and/or ultrastructural analysis of isolated strains or field organisms, have recently led to the description of several new diatom and dinoflagellate species (e.g., Sar et al., 2014; Boutrup et al., 2017; Ferrario et al., 2018; Tillmann et al., 2018; Sunesen et al., 2020). By contrast, little is known about species diversity from other phytoplankton groups that include delicate and small organisms that are hard to identify to species level by microscopy (Silva et al., 2009).
Despite the well-known importance of frontal areas for biomass accumulation in the Argentine Sea, only a few studies have dealt with phytoplankton composition and dynamics in these areas. Most of them have been focused on the shelf-break front (e.g., Carreto et al., 1995; Balch et al., 2014; Carreto et al., 2016), but little is known about tidal fronts, which also play a significant role in biological production and enhanced activity at higher trophic levels (Acha et al., 2004). Among them, the Valdés front has been previously studied because of the occurrence of intense harmful phytoplankton blooms (Carreto et al., 1986). Located in shelf waters around Valdés Peninsula, the Valdés front separates well-mixed and nutrient rich coastal waters from highly stratified and poor nutrient waters offshore. Phytoplankton composition differs at both sides of the front. Chain forming diatoms and dinoflagellates usually dominate at the homogeneous and stratified sides of the front, respectively (Carreto et al., 2007). By contrast, phytoplankton dynamics from other southern frontal areas such as Blanco Cape and de los Estados Island remain almost unexplored (Guerrero et al., 1999; Paparazzo et al., 2010).
Metabarcoding stands as a powerful tool for assessing plankton diversity, particularly in taxonomic groups lacking conspicuous morphological characteristics or with numerous cryptic species (De Vargas et al., 2015). Furthermore, it proves valuable in detecting species that occur at low abundances or are simply beyond the reach of traditional microscopic analyses (Grzebyk et al., 2017). A good correspondence between metabarcoding and microscopy-based abundance and biomass suggests its potential for semiquantitative analysis of plankton, although marked discrepancies have also been noticed (Abad et al., 2016; Gran-Stadniczeñko et al., 2019; Santi et al., 2021; Andersson et al., 2023). Although metabarcoding has been intensively implemented worldwide in the last two decades for phytoplankton analysis (Burki et al., 2021), in the Argentine Sea there is a lack of this type of studies. As the current knowledge we have of the phytoplankton of the Argentine Sea comes mainly from microscopic studies, it is timely to complement both approaches to get a better and more detailed understanding of the phytoplankton community composition in the area.
Here we analyzed for the first-time eukaryotic phytoplankton diversity and community composition in three understudied frontal areas of the Argentine Sea by a combination of 18S metabarcoding and microscopy, considering the strengths and weaknesses of metabarcoding for the study of species richness, dominant taxa, and species assemblages.
2 Methods
2.1 Field sampling and data acquisition
Sampling was carried out in early austral summer on the R/V Bernardo Houssay from January 6th to 12th 2016, along three study areas (Figure 1A) covering a latitudinal gradient: a) Stations 1-2 located in front of Valdés Peninsula (≈43°S), b) Stations 7, 8, 9 located in front of the southern tip of San Jorge Gulf, i.e. Blanco Cape (≈47°S), and c) Stations 15-16 located in front of de los Estados Island (≈55°S). Samples for both metabarcoding and microscopy analyses were simultaneously taken at each sampling station (see below).
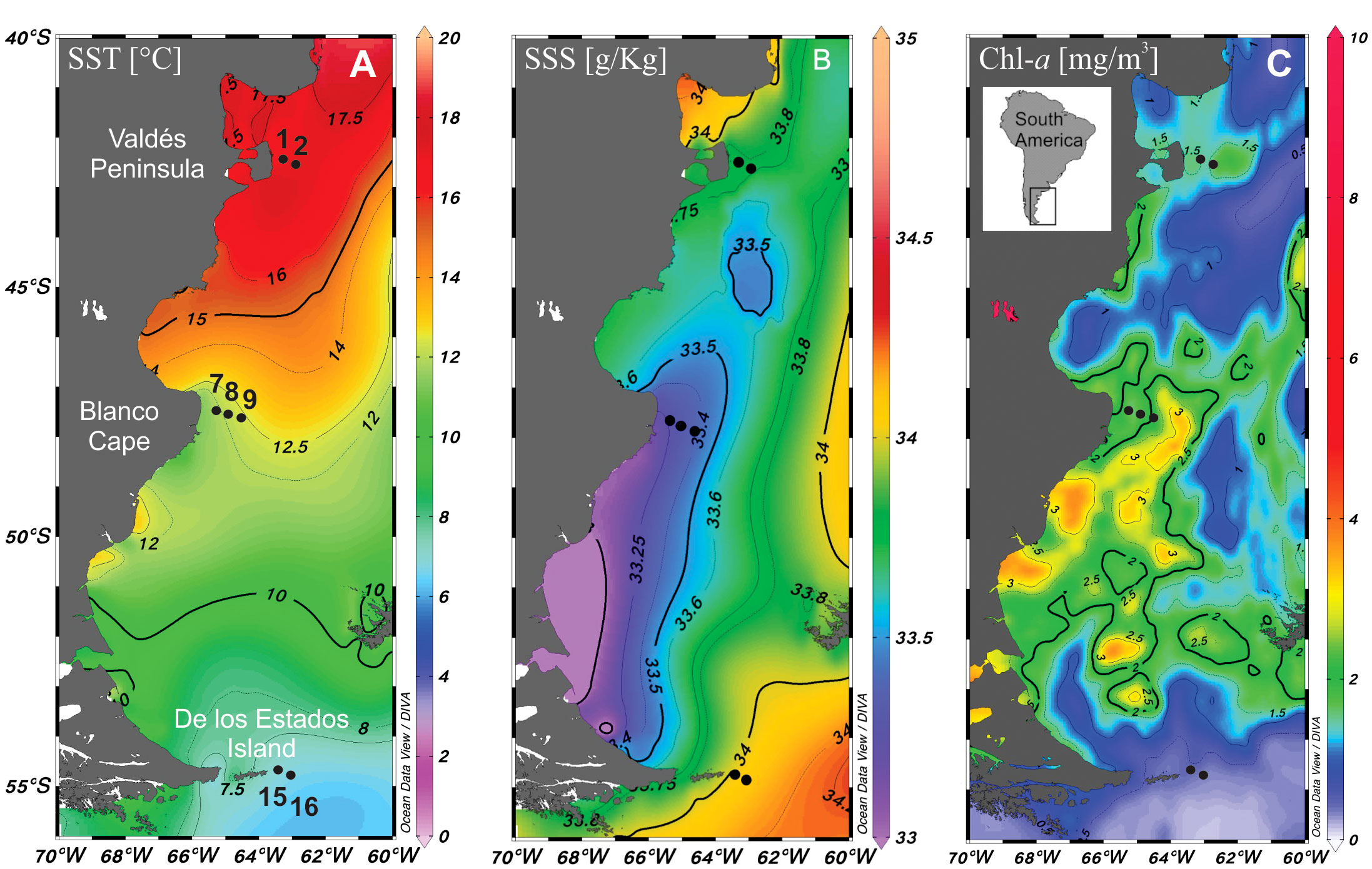
Figure 1 Map of the study area showing the location of sampling stations in the Argentine Sea (black dots). (A) Sea surface temperature (SST), (B) salinity (SSS) and (C) Chl-a distribution.
Physical data on conductivity (salinity)/temperature/depth were determined in situ with a profiling conductivity-temperature-density (CTD) device (Sea Bird SBE 911 plus, Bellevue, WA, USA) for each sampling station. Moreover, the World Ocean Atlas (WOA-18) from the NOAA/National Center for Environmental Information website (https://www.nodc.noaa.gov/OC5/woa18/) was used to download January 2016 gridded mean fields of Sea Surface Temperature (Locarnini et al., 2019) and Sea Surface Salinity (Boyer et al., 2018; Zweng et al., 2018). Satellite chlorophyll a (Chl-a) concentration was taken from https://neo.gsfc.nasa.gov/view.php?datasetId=MY1DMW_CHLORA&year=2016. All data were plotted with ODV software version 5.6.3 (odv.awi.de).
Surface (5 m) seawater samples of 4-5 L were collected at each station with Niskin bottles. Aliquots of 250 mL were fixed with acidic Lugol’s iodine solution (4%) for microscopic analysis of phytoplankton composition. The rest was sieved through a 200 µm Nitex mesh to remove large zooplankton. Aliquots of 2 L were then sequentially filtered through 5 µm and 0.4 µm pore polycarbonate filters for molecular analysis. These filters were bent and wrapped into foil, flash frozen in liquid nitrogen and kept at -20°C for 4-10 days. The upper surface of the filters was then completely scraped using a 2 ml lysis buffer and a sterile scraper. The collected fluid was stored in Eppendorf tubes and frozen at -20°C until DNA extraction.
2.2 Molecular analysis
Genomic DNA was isolated from the filters using the Nucleospin Plant II kit (Machery-Nagel, Düren, Germany) according to the manufacturer’s protocol. DNA concentrations were determined with a Quantus Fluorometer according to the manufacturer’s protocol for measuring double stranded DNA (QuantiFluor dsDNA System, Promega, USA).
The variable V4 region of the 18S rDNA was amplified using the following primer-set: 528iF (GCG GTA ATT CCA GCT CCA A) and 964iR (AC TTT CGT TCT TGA TYR R) (Metfies et al., 2020). The PCR reaction mixture consisted of 12.5 μL master mix (KAPA HiFi HotStart ReadyMix, KAPABiosystems, Roche), 5 μL of each primer (1 μM) and 2.5 μL template (~5 ng/μL). Amplification was carried out on a MasterCycler (Eppendorf, Hamburg, Germany) with the following program: 95°C for 3 min, 25 cycles consisting of: 95°C for 30 s, 55°C for 30 s and 72°C for 30 s. Followed by a final five minutes at 72°C. The products were kept at 4°C once the program had ended. Samples were stored at −20°C until further analysis. The PCR products were purified from an agarose gel 1% [w/v] with the AMPure XP PCR purification kit (Beckman Coulter, Ing., USA) according to the manufacture’s protocol. After purification of the 18S rDNA fragment the DNA concentrations of the samples were determined using the Quantus Fluorometer (Promega, USA). Indices and sequencing adapters of the Nextera XT Index Kit (Illumina, USA) were attached via the Index PCR. All PCRs had a final volume of 50 μl and contained 25 μl of KAPA HIFI Mix (Kapa Biosystems, Roche, Germany), 5 μl of each Nextera XT Index Primer [1 μmol/L], 5 μl DNA-template [~5ng] and 10 μl PCR grade water. PCR amplification was performed in a thermal cycler (Eppendorf, Germany) with an initial denaturation (95°C, 3 min) followed by 8 cycles of denaturation (95°C, 30 sec), annealing (55°C, 30 sec), and extension (72°C, 30 sec) with a single final extension (72°C, 5 min). Prior to quantification of the PCR products with the Quantus Fluorometer (Promega, USA) for sequencing with the MiSeq-Sequencer (Illumina, USA), the final library was cleaned up using the AMPure XP PCR purification kit (Beckman Coulter, Ing., USA). Sequencing of the DNA-fragments was carried out with the MiSeq Reagent Kit V3 (2 x 300 bp) according to the manufacturer’s protocol (Illumina, USA).
Raw reads were quality trimmed with Trimmomatic (Bolger et al., 2014). Thus, reads were scanned with a 4-base wide sliding window and cut when the average quality dropped below 15. For merging paired-end reads, the script join-paired-ends within the open-source bioinformatics pipeline QIIME v.1.8.0 was used with a minimum read overlap of 20 bases. Further analyses were performed following a pipeline developed in-house using QIIME v.1.8.0 (Caporaso et al., 2010). In short, reads were quality-filtered according to recommended settings (Bokulich et al., 2013). Only sequences that fully matched the primer sequences at the beginning and end of the sequence, respectively, and which were between 200 and 500 bp in length were further processed. For chimera detection and clustering of sequences into OTUs we used the QIIME workflow ‘usearch.qf’, which incorporates UCHIME (Edgar et al., 2011). Pre-clustered sequences were checked for chimeras (de novo and with Silva 119 SSU). The remaining sequence set was clustered (de novo) into OTUs with a similarity threshold of 98%. Sequences from all samples were analyzed together to define operational taxonomic units (OTUs) for 18S rRNA genes using SWARM (Mahé et al., 2015), with a cut-off value of 0,005% of the total amount of reads. OTUs were assigned to taxa based on their similarity with SILVA database. In addition, BLAST searches were performed to further explore identities of the most abundant OTUs.
The database initially contained a set of 3,460 OTUs. After applying a cut-off value of 0,005% and selecting those assigned to Chlorophyta, Haptophyta, Cryptophyta, Stramenopiles (excluding Phaeophyceae, Labyrinthulomycetes, and Peronosporomycetes), Dinophyta, Raphidophyceae, Picozoa, and Telonemians, the number of OTUs was reduced to 382.
We rarefied all samples to 43,456 reads, the smallest phytoplankton reading determined by the rarefaction curves (Cameron et al., 2021). Rarefaction curves were created and calculated using the R package vegan (Oksanen, 2015).
2.3 Microscopic analysis
The abundance of nano- (5-20 µm) and microplankton (20-200 µm) was estimated following the Utermöhl (1958) method, with a phase-contrast Leica DMIL LED equipped with a Mshot MS60 camera. Subsamples (50 mL) from the water obtained by Niskin bottles were left to settle for 24 h in a composite sedimentation chamber before cell counting. At least 300 cells of the dominant taxa were counted in random fields or in half the chamber to estimate general phytoplankton composition, whereas the whole chamber bottom was scanned to count less abundant species. All taxa, including photosynthetic and heterotrophic groups, were identified to the highest possible taxonomic level (i.e., species). However, since flagellates generally lose their flagella by the addition of fixatives, unidentified phytoflagellates and round-shaped organisms with or without flagella, were included in a single group as small unidentified flagellates (< 10 µm), referred herein as small flagellates. Further morphological examination was conducted by phase-contrast, differential interference contrast and UV epifluorescence microscopy with a Leica DM2500 microscope equipped with a DFC420C camera. For the estimation of phytoplankton biomass, we utilized cell carbon content (pg C L-1) data from the most abundant taxa, obtained from previous studies conducted in the area by the authors (e.g., Almandoz et al., 2011).
2.4 Data analysis
Cluster analyses based on Euclidean distances and Unweighted Pair Group Average (UPGMA) algorithm were carried out with the software PRIMER v6. Relative biomass values of taxa representing at least 0.5 of total biomass (N=23) and normalized OTU values (N=382) were used. The Similarity Profile routine (SIMPROF) test was further applied for detecting significant clusters of samples.
3 Results
3.1 Hydrography
The three studied zones differ in their sea surface temperature and salinity, with warmer (≈16°C) and saline (33.8) waters at Valdés Peninsula, colder (≈8°C) and saline (34) waters at de los Estados Island, and waters with intermediate temperature (≈12°C) and lower salinity (33.4) at Blanco Cape (Figures 1B, C). Satellite Chl-a estimations showed higher values (≈2 mg m-3) in the two northern frontal areas, and lower values to the south (Figure 1C). Vertical profiles showed a typical tidal front structure at Valdés Peninsula and Blanco Cape zones, with well-mixed waters near shore and enhanced stratification offshore (Figure 2). By contrast, a topographically controlled front is observed at de los Estados Island, when the low salinity water coming from the Beagle Channel converges with the cool and salty water from the upper stratum of the Malvinas Current (Figure 2), generating a surface front at the edge of the continental slope (Guerrero et al., 1999).
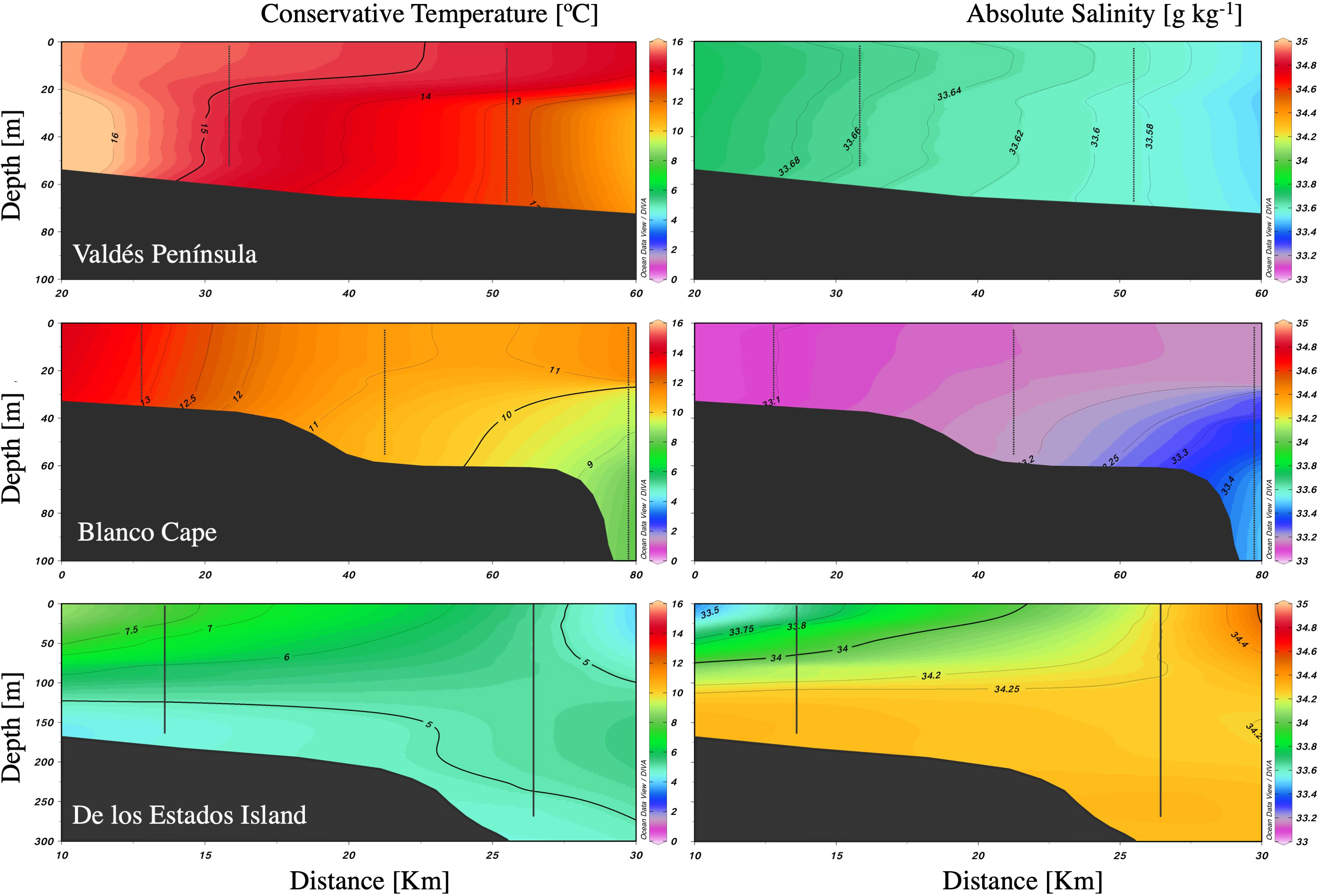
Figure 2 Vertical profiles of temperature and salinity along three frontal areas in the Argentine Sea.
3.2 Overall phytoplankton composition
Microscopy cell counts revealed that phytoplankton was mainly composed of small flagellates in terms of cell abundance, reaching up to 11.6 × 106 cells L-1 at Valdés Peninsula (St. 2), that represented 90.6% of total cells (Figure 3A). Small flagellates were only outnumbered by diatoms at de los Estados Island, where diatoms reached 7.0 × 105 cells L-1 (St. 16), accounting for 56.4% of total cell abundance (Figure 3A). Other taxonomic groups, i.e., dinoflagellates, prasinophytes, haptophytes, raphidophytes, and silicoflagellates were detected in low values (< 7%), except for cryptophytes which accounted for up to 15.2% of total cells at Blanco Cape (St. 9).
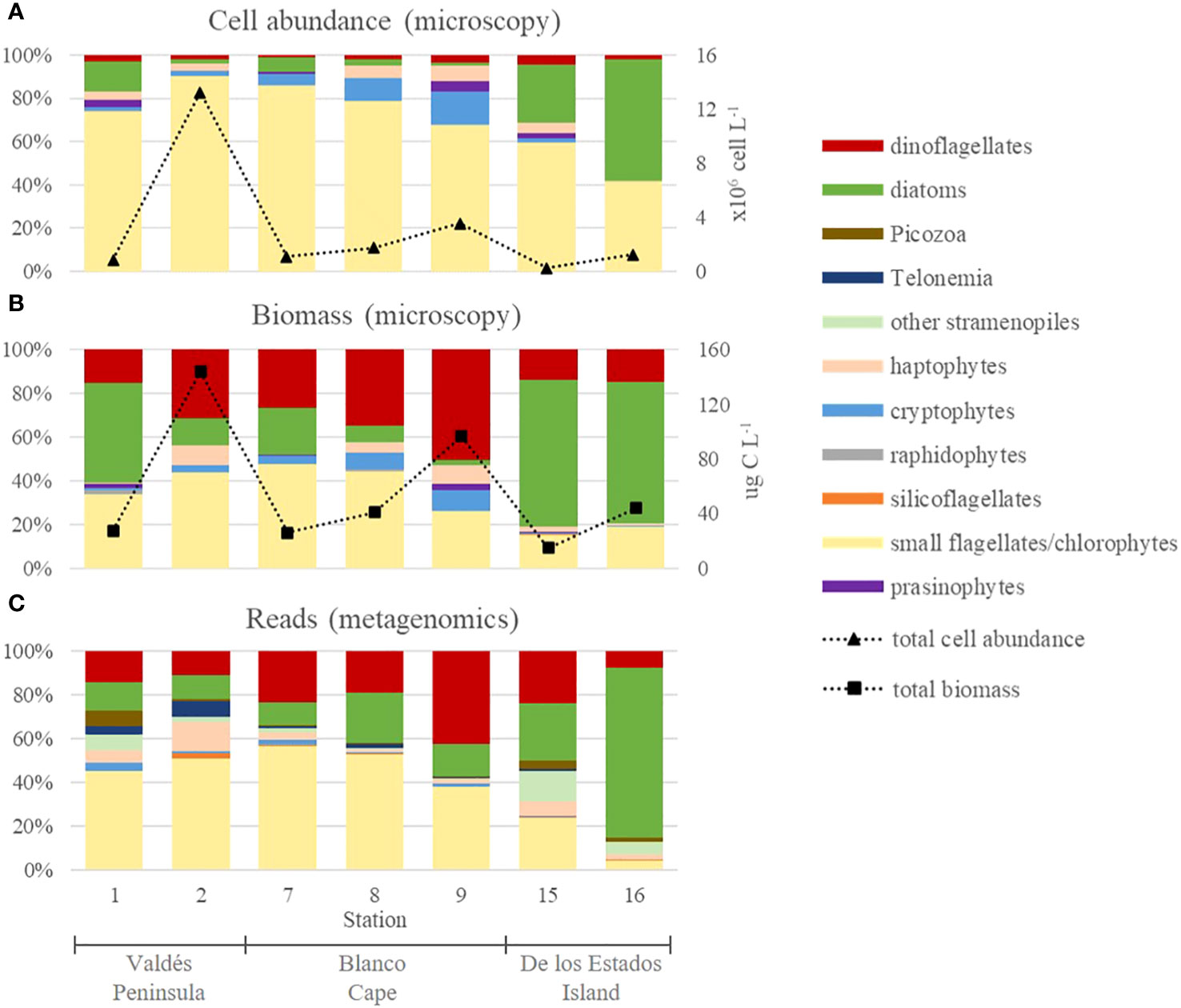
Figure 3 Overall phytoplankton composition in frontal areas of the Argentine Sea expressed in relative cell abundances (A) and relative carbon biomass (B) of main phytoplankton groups obtained by microscopy, and relative reads of main phytoplankton groups obtained by metabarcoding (C). Note that small flagellates that could not be identified by microscopy were mostly assigned to chlorophytes based on metabarcoding analysis.
Carbon biomass estimations, based on cell volumes, showed conspicuous differences with respect to cell abundance distribution (Figure 3B). Because of their bigger cell size, dinoflagellates dominated at Valdés Peninsula (St. 9), reaching up to 48.7 µg C L-1 (50.3% of total biomass), whereas diatoms mainly dominated at de los Estados Island, reaching up to 28.3 µg C L-1 (64.7% of total biomass) at St. 16 (Figure 3B). Small flagellates reached a maximum of 62.8 µg C L-1 (43.6% of total biomass) at St. 2. The other taxonomic groups represented less than 10% in all samples.
Metabarcoding analysis showed that total reads were dominated by chlorophytes, dinoflagellates, and diatoms (Figure 3C), in accordance with what was observed for microscopic biomass estimations. Other groups, such as cryptophytes, silicoflagellates, raphidophytes, and haptophytes were detected in low relative read numbers. Moreover, Telonemians, Picozoa, and other non-diatom Stramenopiles, which could not be identified by microscopy, were also detected in low read numbers. Chlorophytes dominated at Valdés Peninsula and at St. 7-8 from Blanco Cape, where they represented 52%-57% of total reads, similar to the dominance of small flagellates observed by microscopy. This observation suggests that most cells identified during cell counting as small flagellates in these samples may correspond to chlorophytes. Chlorophytes were mostly detected in the 0.4 µm filters than in 5 µm filters (Supplementary Figure 1A), reaching up to 97.3% of total reads (St. 8). By contrast, dinoflagellates and diatoms were more important in the 5 µm filters (Supplementary Figure 1B), reaching up to 70.3% (St. 9) and 93.5%, (St. 16), respectively.
3.3 Species richness
Microscopy analysis revealed the presence of 91 taxa, most of them diatoms (40) and dinoflagellates (40), (Figure 4). The richest taxa include dinoflagellates from the genus Protoperidinium (8), Ceratium (4), and Prorocentrum (3), and the diatoms Pseudo-nitzschia (5), Guinardia (3), and Thalassiosira (3) (Supplementary Table 1). By contrast, the other groups were mostly assigned to class level.
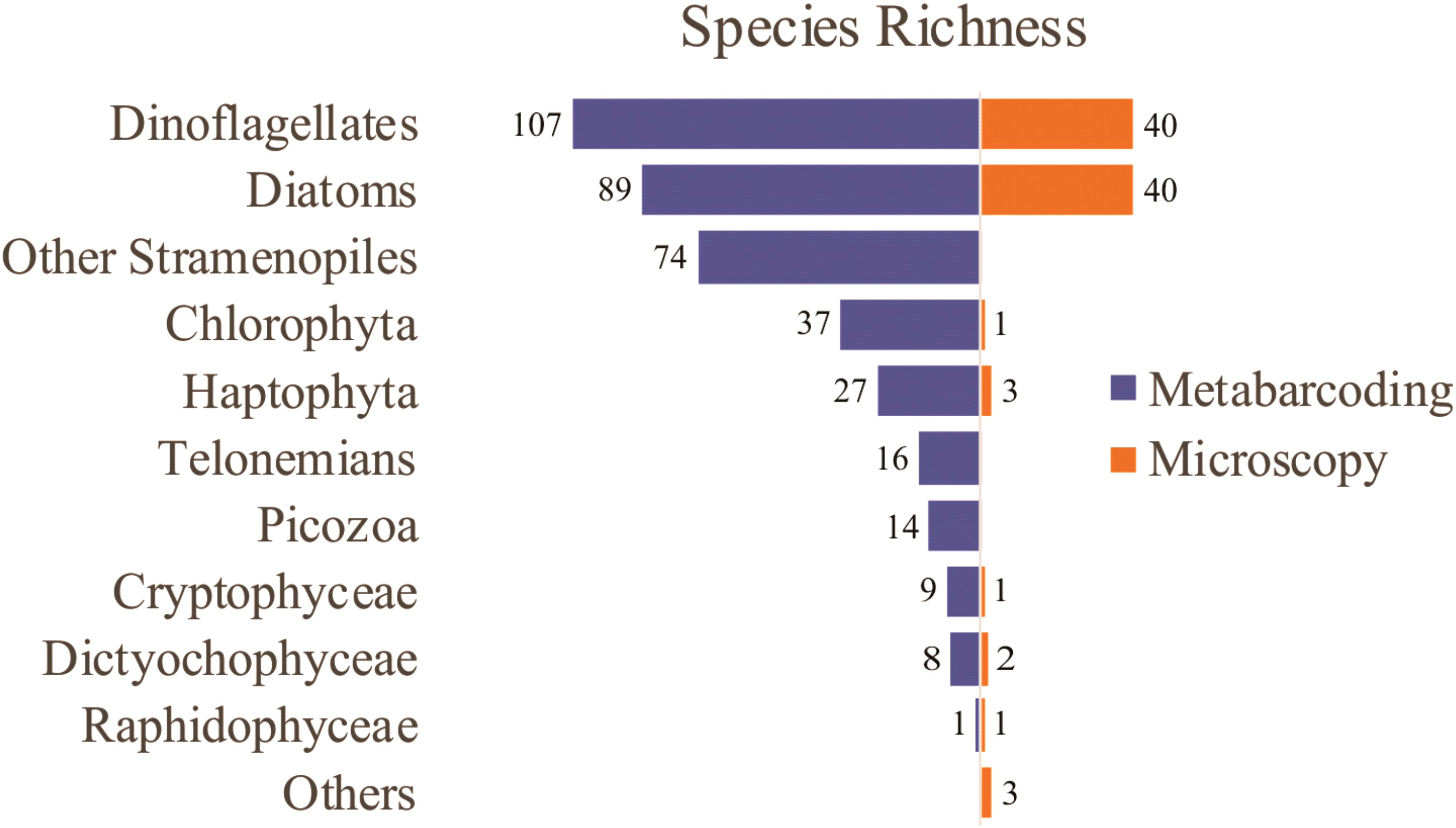
Figure 4 Phytoplankton species richness detected by microscopy and metabarcoding in sampling sites from the Argentine Sea.
Metabarcoding analysis revealed a notably higher number of taxa, with a total of 382 environmental OTUs, most of them assigned to dinoflagellates (107), diatoms (89), and other Stramenopiles (74), followed by Chlorophyta (37), Haptophyta (27), Telonemians (16), Picozoa (14), Cryptophyceae (9), Dictyochophyceae (8), and Raphidophyceae (1) (Figure 4). The richest taxa include the diatoms Chaetoceros (21), Thalassiosira (16), Pseudo-nitzschia (8), and Guinardia (7), the dinoflagellates Gyrodinium (10), the Gymnodinium clade (8), and Symbiodinium (7), other Stramenopiles such as uncultivated MArine STramenopiles (MAST) linages (10) and Triparma (5), the Chlorophyta genus Micromonas (6), and the Haptophyta genera Chrysochromulina (6) and Phaeocystis (4) (Supplementary Table 2).
3.4 Species assemblages and dominant taxa
Cluster analysis based on relative biomass estimations (microscopy) showed three main groups (Figure 5A). Stations from Blanco Cape (St. 7-9) grouped together with the offshore station from Valdés front (St. 2) and were characterized by the presence of an assemblage dominated by small flagellates, which accounted for up to 44.2% of total biomass at St. 2 (Supplementary Table 3). In terms of relative biomass, small flagellates were followed, at St. 2 by the dinoflagellate aff. Peridiniella/Scripsiella (21.5%) and unidentified small centric diatoms (10.7%), and at St. 7 by the diatom Guinardia delicatula (10.1%), and the dinoflagellates Protoperidinium granii (9.2%) and P. conicum (8.5%). A high contribution of Gyrodinium sp. (≈ 30 µm) was observed at stations 8-9, with increased cell abundance (9.5 × 104 cells L-1) and biomass contribution (46.3%) towards offshore waters (St. 9). Cryptophytes were also observed in high densities at stations 8-9, with cell densities up to 5.4 × 105 cells L-1 and biomass contribution of 9.6% at St. 9 (Figures 3A, B). A second group was formed with samples from de los Estados Island (St. 15-16), here diatoms outnumbered small flagellates in terms of relative biomass. Stephanophyxis nipponica (26.4%) and Cerataulina pelagica (22.7%) were the most important diatoms at St. 15, whereas Pseudo-nitzschia spp. dominated at St. 16 (60.9%), reaching 4.6 × 105 cells L-1. Other diatom species, such as Fragilariopsis kerguelensis, Pseudo-nitzschia turgiduloides, and single cells resembling Attheya septentrionalis (Figure 6), were also observed by microscopy at St. 16. Finally, St. 1 did not group with any other stations, presented an assemblage dominated by the diatom Asterionellopsis glacialis (47.5%), followed by small flagellates (37.5%) and the dinoflagellate Prorocentrum micans (10.6%). The most abundant taxa found in the three frontal areas by microscopy are illustrated in Figure 6.
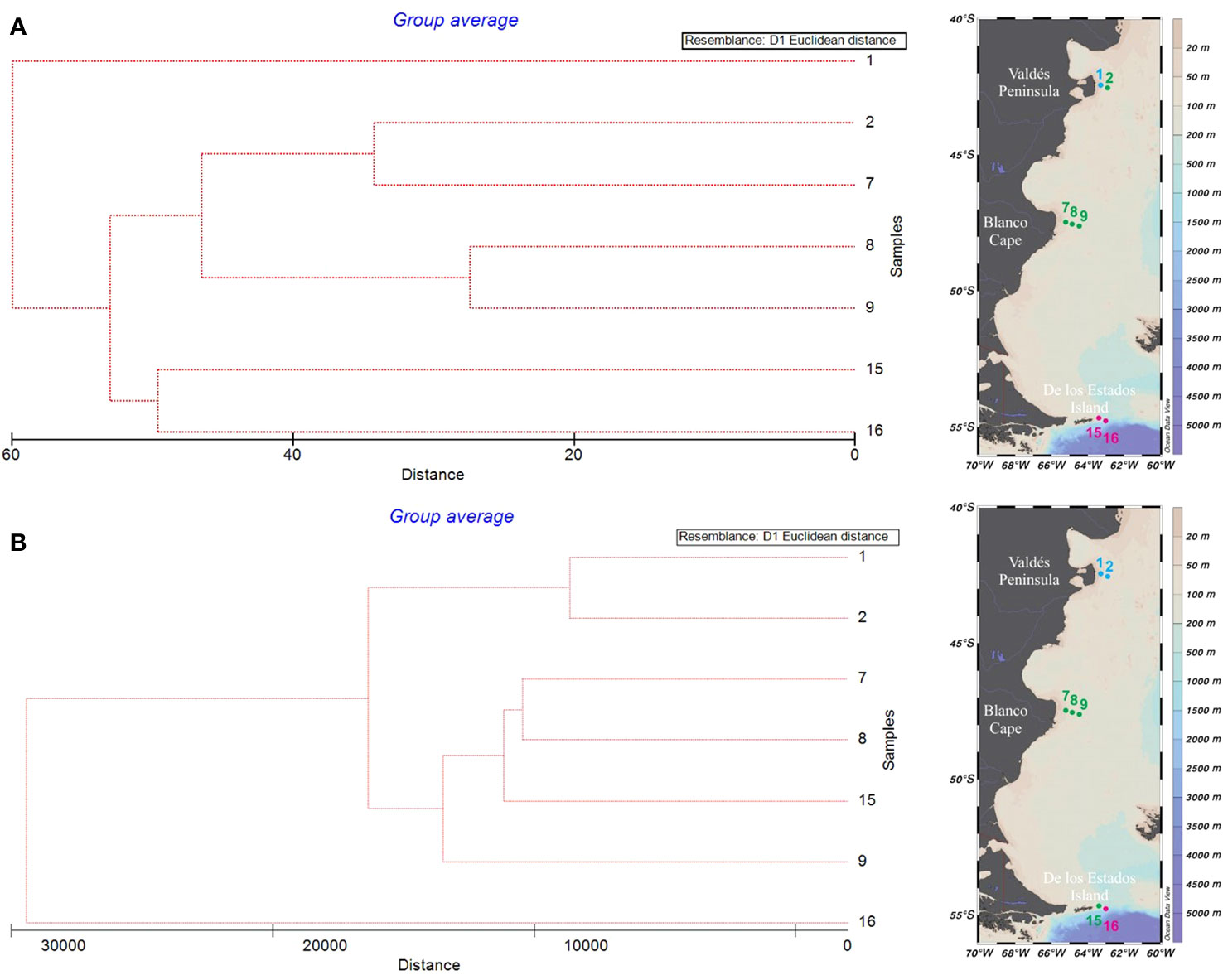
Figure 5 Cluster analysis based on Euclidean distance and Unweighted Pair Group Average (UPGMA) algorithm of main phytoplankton taxa relative biomass identified by microscopy (A), and relative read numbers obtained by metabarcoding (B) in samples from the Argentine Sea.
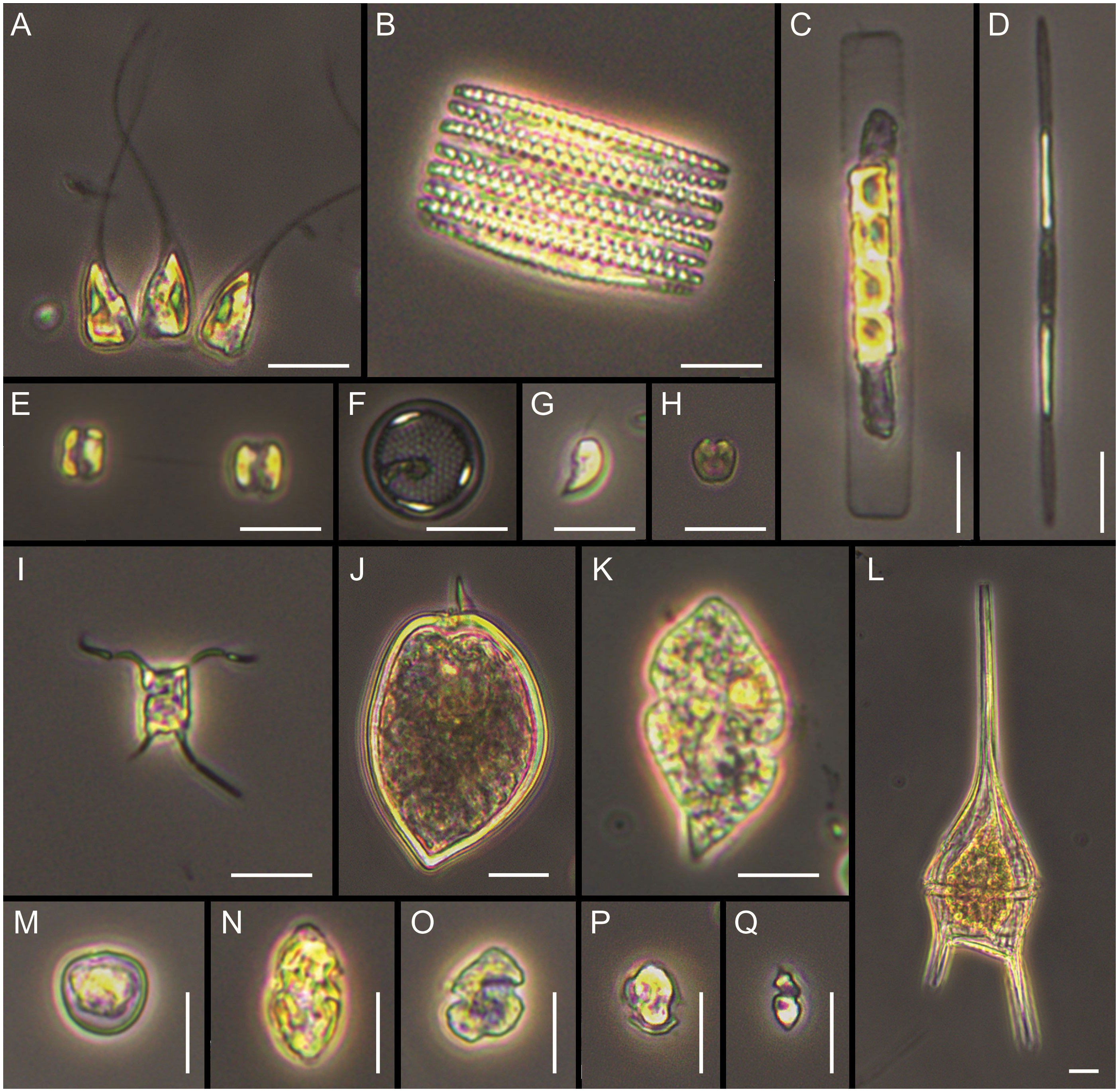
Figure 6 Main taxa observed under light microscopy. (A) Asterionellopsis glacialis, (B) Fragilariopsis kerguelensis, (C) Guinardia delicatula, (D) Pseudo-nitzschia aff. turgiduloides, (E, F) unidentified small centric diatoms, (G) unidentified cryptophyte, (H) unidentified prasinophyte, (I) Attheya aff. septentrionalis, (J) Prorocentrum micans, (K) Gyrodinium sp., (L) Tripos lineatus, (M) Prorocentrum cordatum, (N, O) gymnodinoid dinoflagellates, (P) small thecate dinoflagellate, (Q) small naked dinoflagellate. Scale bars = 10 µm.
Cluster analysis based on metabarcoding also showed three main groups that had some differences with the groups previously found by microscopy (Figure 5B). An assemblage mainly represented by the prasinophyte species Pycnococcus provasolii was observed at St. 1-2 at Valdés front (Supplementary Table 4). Pycnococcus provasolii accounted for 54.7% and 85.4% of total Chlorophyta reads at St. 1 and 2, respectively. Other species with high contribution at Valdés front included Phaeocystis globosa (Prymnesiophyceae), Chloropicon roscoffensis (Prasinophytes, Clade VIII), Picomonas judraskeda (Picozoa), the dinoflagellate Prorocentrum micans, an unclassified Dinophyceae, and the diatom Thalassiosira profunda. In contrast with the large contribution to total biomass of the diatom Asterionellopsis glacialis at St. 1 (47.5%) detected by microscopy, this species accounted for less than 0.2% of total reads. A second group was formed by samples from Blanco Cape (St. 7-9), and the inner station from de los Estados Island (St. 15). Here the assemblage was mainly represented by OTUs assigned to the Clade VIII (subclade B) of prasinophytes, Picochlorum sp. (Trebouxiophyceae), the diatom Minidiscus trioculatus, and the dinoflagellate Gyrodinium fusiforme. Finally, the offshore station from the los Estados Island (St. 16) did not group with any other sample, here the assemblage was dominated by the diatom species Chaetoceros contortus, accounting for 63.8% of total reads. This observation markedly differs with what we found by microscopy at St. 16, where total biomass was dominated by Pseudo-nitzschia spp. (60.9%), and Chaetoceros spp. contributed less than 5%.
4 Discussion
4.1 Main phytoplankton groups
Microscopy and 18S metabarcoding both identified small flagellates, dinoflagellates and diatoms to be the dominant taxonomic groups within the three different frontal areas sampled along the Argentine Sea. Here, 18S metabarcoding provided a more detailed insight into the taxonomic composition of the small flagellates, suggesting small chlorophytes to be main contributors to this taxonomically unidentified group. In combination, the two methods indicate a prevalence in the predominance of small chlorophytes at Valdés Front, an increase in dinoflagellates in offshore waters near Blanco Cape, and a dominance of diatoms at de los Estados Island. More particular, we observed agreement on the main taxonomic groups when comparing metabarcoding reads and carbon biomass measurements in samples with high biomass (Figure 3, St. 2, 9, and 16). As expected, there was less agreement between metabarcoding reads and cell abundance. Notably, cell abundance did not reflect the increased presence of dinoflagellates in the waters near Blanco Cape. In this context, our findings align with recent previous research that suggest a stronger correlation between metabarcoding reads and carbon biomass compared to cell abundance in microbial communities (Andersson et al., 2023). This correlation may be attributed to a positive relationship between the genome size of eukaryotes (and the number of rDNA copies) and their cell size, as highlighted by Lavrinienko et al. (2021).
4.2 Spatial variability of main species assemblages
Cluster analyses performed on both relative reads (metabarcoding) and relative biomass (microscopy), grouped the samples into three main groups well related to the geographic location of the sampling stations. Stations 7-9 (Blanco Cape) clustered closely, while the most norther station 1 (Valdés Peninsula) and the most southern station 16 (De los Estados Island) clustered both separately outside the Blanco Cape cluster. Differences in the clustering were observed between the methods for stations 2 and 15. Station 2 clustered with station 1 based on the metabarcoding data, but with stations 7-9 based on the microscopic data, while Station 15 clustered with station 16 based on the microscopic data and with stations 7-9 based on the molecular data. We consider that these differences were largely due to the higher taxonomic resolution provided by metabarcoding, particularly of small flagellates identified as a single group by microscopy, and to differences in the detection of dominant diatoms by both methods (St. 1 and 16). Microscopy separated St. 1, north of Valdés Peninsula, from the rest of the stations mainly because of the high relative biomass of the diatom Asterionellopsis glacialis, which was not reflected in terms of relative reads. This is a major bloom-forming species in coastal areas worldwide, for which high cryptic diversity has recently been observed (Kaczmarska et al., 2014). The low values of relative reads corresponding to Asterionellopsis could be associated, among other factors, to its particularity of producing extracellular mucous-like compounds, which has been observed to interfere with DNA amplification (Odebrecht et al., 2013; Uyua et al., 2014). However, we have no way of testing whether these compounds were indeed present and if so whether they affected other species in the sample as well as Asterionellopsis causing a reduction in total reads. In any case, this is something we cannot determine due to the compositional nature of metabarcoding, which prevents us from comparing the numbers of total reads obtained in different samples (Gloor et al., 2017). Moreover, our metabarcoding data suggest that small flagellated prasinophyte species could represent a widespread and important component of phytoplankton assemblages in the Argentine Sea during late spring or summer, further contributing to the differences in the clustering observed between the methods. An important bloom of the chlorophyte species Pycnococcus provasolii was observed on the stratified side of Valdes Peninsula, in accordance with microscopic observations. This is a little known, small (≈1.5–4 μm) and coccoid species described based on cell cultures from the northwestern Atlantic and the Gulf of Mexico (Guillard et al., 1991), which has been tentatively identified by transmission electron microscopy in the Strait of Magellan (≈52°-54°S) dominating phytoplankton during summer (Zingone et al., 2011). Likewise, pigments mostly associated with chlorophytes in coincidence with a massive bloom of up to 59.1 × 106 cells L-1 of a picoplanktonic (<3 μm) coccal cells of an unidentified chlorophyte, were detected in the shelf-break frontal area of the Argentine Sea during late spring (Carreto et al., 2008). Moreover, a small coccal cell (3 μm) possibly belonging to the group of prasinophytes was an important component of phytoplankton assemblages in stratified waters north of 52°S during a summer oceanographic cruise in the southern Argentine Sea (Antacli et al., 2018). Thus, we consider the Pycnococcus provasolii bloom detected during January in our study could represent a later stage of phytoplankton succession at Valdés Front area.
Based on both methods, the frontal area of Blanco Cape presented the typical phytoplankton composition observed in local tidal fronts (Carreto et al., 2007; Paparazzo et al., 2010), with a predominance of diatoms and dinoflagellates in coastal (mixed) and offshore (stratified) waters, respectively. Vertical mixing in the coastal side of the front brings bottom nutrients up to the upper layers, which promotes the dominance of diatoms. By contrast, dinoflagellates dominate in nutrient-depleted and stratified waters offshore (Ramond et al., 2021). Dinoflagellates reads were particularly abundant in the more external station, where the species Gyrodinium fusiforme represented 88.2% of total dinoflagellates reads and a peak cell abundance of 9.5 × 104 cells L-1 of Gyrodinium sp. was observed by microscopy. However, the dominant Gyrodinium cells observed with microscopy did not match completely with the description of G. fusiforme (Takano & Horiguchi, 2004) as they were smaller (≈30 μm long) and had a more rounded epicone. Likewise, smaller Gyrodinium cells recently observed in shelf-break waters of the Argentine Sea, could not be assigned to species level by detailed microscopy, due to their differences with other described Gyrodinium species, and probably represents a new species (Fabro & Almandoz, 2021). Previous phytoplankton studies at Valdés Front area revealed the occurrence of a conspicuous dinoflagellate bloom dominated by the toxic species A. catenella causing water discoloration from the beginning of October until January, with cell densities up to 1.8 × 106 cells L-1 during November 1981 in the stratified side of the front (Carreto et al., 1986; Carreto et al., 2007).
Stations at de los Estados Island were also grouped differently, microscopy results grouped together stations 15 and 16, based on the high contribution of diatom species to total biomass, mainly Stephanophyxis nipponica and Cerataulina pelagica at. St. 15 and Pseudo-nitzschia spp. at St. 16. By contrast, metabarcoding results showed that St. 15 presented a composition similar to that of Blanco Cape, with high relative reads of an uncultured unidentified eucaryote, Pycnococcus provasolii, Minidiscus trioculatus, and Gyrodinium fusiforme whereas offshore waters (St. 16) were dominated by the diatom species Chaetoceros contortus. The differences in the clustering between the methods are probably related to missing sequence information for the 18S gene of Stephanophyxis nipponica in the reference data-base (Santoferrara, 2019). Beyond this, we could not find a clear explanation for the discrepancy observed in the identification of dominant species at St. 16 by microscopy (Pseudo-nitzschia spp.) and metabarcoding (C. contortus). Discrepancies in the identification of dominant taxa between microscopy and metabarcoding have been previously documented in the scientific literature (Abad et al., 2016; Mäki et al., 2017; Hanžek et al., 2023). These disparities can often be attributed to multiple factors, including potential error stemming from various stages of the analysis process. These factors encompass sample preservation, the volume of the sample analyzed, the DNA extraction process, primers selection, PCR procedures, incomplete reference databases, data analysis inaccuracies, and others relevant variables (Santoferrara, 2019; Burki et al., 2021; Santi et al., 2021). Particularly for the genus Pseudo-nitzschia, Ruggiero et al. (2015) found an under-estimation of its relative proportion of metabarcoding reads compared with microscopy observations in the Gulf of Naples. By contrast, Turk Dermastia et al. (2023) found a large proportion of Pseudo-nitzschia by 18S-v9 and rbcL metabarcoding that was not observed by microscopy in the Gulf of Trieste, mainly attributed to misidentification of small and delicate species by microscopy. Therefore, our results show that there could be notable differences in the identification of dominant organisms at species level with microscopy and metabarcoding, which could lead to different ecological interpretations. This highlights the need for more comparative studies to attempt to bridge this gap, but on the other hand our results demonstrate that both methodologies are currently complementary (Pierella Karlusich et al., 2020).
Samples from de los Estados Island are influenced by a topographically controlled front with strong horizontal salinity and temperature gradients as a result of the mix of the low salinity water coming from the Beagle Channel with the cool and salty water from the upper stratum of the Malvinas Current (Guerrero et al., 1999). Consequently, a stratified water column was found in shallow coastal waters and a mixed water column was observed offshore. In a previous phytoplankton study covering a transect from the Beagle Channel to the Burdwood Bank, high cell densities of Chaetoceros spp. and Pseudo-nitzschia spp. were also recorded to the east of de los Estados Island during December, associated with high nutrients and low light conditions (Guinder et al., 2020). The high contribution of diatoms and the different species composition found between inshore and offshore waters in de los Estados Island frontal area, could be then associated with the presence of diatom-rich Antarctic waters moving northward through the Malvinas current (Guerrero et al., 1999).
4.3 Phytoplankton richness in the Argentine Sea
One of the most conspicuous feature of phytoplankton composition in the study area with the high dinoflagellate richness, compared with other groups. This observation agrees with the Tara Ocean global survey, which has shown that overall dinoflagellate 18S richness accounted for ∼49% of total protistan richness (Le Bescot et al., 2016). Dinoflagellate diversity assessments in the Argentine Sea have been previously confined to microscopic observations showing also a high morphological diversity (e.g., Balech, 1988; Balech, 2002). In a more recent study of summer phytoplankton diversity in the southern Argentine Sea (47°–55°S), dinoflagellates represented 36% of total species/morpho-species detected by microscopy (Antacli et al., 2018). According to these authors, only 46 of 148 dinoflagellate taxa could be identified to genus or species level, most of them corresponding to the genus Protoperidinium. Our summer metabarcoding analysis showed a total of 107 OTUs assigned to dinoflagellates, from which a large number was assigned to Gyrodinium, the Gymnodinium clade, and Symbiodinium, but also a high percentage was assigned to unclassified Dinophyceae and Peridiniphycidae. By contrast, only one OTU was assigned to the genus Protoperidinium, which contrasts with microscopic observations where at least eight different species were detected in very low cell abundances. Molecular identification of Protoperidinium species is limited by the low number of available sequences in databases, because of their heterotrophic conditions that difficult their culture and growth (Phan-Tan et al., 2016). Up to ten dinoflagellate genera detected by metabarcoding in the present study (see below) have not been mentioned before in the Argentine Sea by previous microscopic observations. They mostly include small size (< 15 µm), with a very thin theca or naked organisms, which require detailed ultrastructural observations for their identification. Moreover, some of them also show complex life cycles involving parasite stages or long cyst stages, reducing the planktonic motile stage, so they could be overlooked in routine monitoring or abundance estimations. This is the case of the genera Symbiodinium, Gymnoxanthella, Chytriodinium, Paragymnodinium, Pfiesteria, Crypthecodinium, Biecheleria, and Pelagodinium, which might be considered in this study as “small naked dinoflagellates’’ or included as Gymnodinium sp. by microscopic analysis (Bhaud et al., 1991; Burkholder et al., 2001; Gómez et al., 2009; Moestrup et al., 2009; Kang et al., 2010; Siano et al., 2010; Yuasa et al., 2016; Decelle et al., 2018). Conversely, the genera Gotoius and Sinophysis consist of large thecate dinoflagellates, with cell sizes exceeding 30 µm and distinct morphological characteristics such as cell shape and theca plate pattern. These characteristics make their identification considerably more straightforward and reliable when employing optical methods (Dodge and Toriumi, 1993; Hoppenrath, 2000). However, it is noteworthy that these genera have not been previously documented in the Argentine Sea, which suggests that they may exist in exceedingly low abundances, consistent with their limited relative read numbers. In this sense, it is important to consider that the significantly larger sample volume used for metabarcoding (4-5 L) in comparison to microscopy (50 mL) could have play a pivotal role in enabling the detection of rare species. These species, although present might have eluded observation under the light microscope techniques.
Diatoms represented the second group in terms of phytoplankton richness, with most OTUs assigned to the genera Chaetoceros, Thalassiosira, and Pseudo-nitzschia. These are among the most diverse marine diatom genus worldwide and have also shown a high species diversity by detailed microscopy studies in the Argentine Sea (Ferrario and Sar, 1988; Sar et al., 2002; Sunesen et al., 2008; Almandoz et al., 2017). By contrast, the finding of the genus Plagiostriata in the metabarcoding analysis represents, as far as we know, a new record for the Argentine Sea. Non-diatom stramenopiles richness was mostly composed of MAST, a group of world-wide distributed marine heterotrophic picoeukaryotes that are mostly phagotrophic free-living flagellates (Massana et al., 2004). Likewise, five OTUs were assigned to the picophytoplanktonic genus Triparma (ex Bolidomonas), which to the present day only includes two described species (Guillou et al., 1999), suggesting this group might have a greater diversity.
Other poorly studied taxonomic groups of nano- and picoplankton showed a great diversity previously ignored in the Argentine Sea. A high OTU richness was found for the heterotrophic pico- and nano-eukaryotes genera Picomonas (5) and Telonema (16), members of two new recently described eukaryotic phyla (i.e., Picozoa and Telonemia) with uncertain evolutionary position and unknown diversity (Shalchian-Tabrizi et al., 2007; Seenivasan et al., 2013). Chlorophytes richness was mainly represented by the genera Micromonas and Prasinoderma, which includes tiny (< 3 µm) flagellates (Micromonas) and nanoplanktonic (≈3-5 µm) coccoid organisms (Prasinoderma), which are hardly detected during microscopic observations (Hasegawa et al., 1996; Guiry, 2022). Moreover, two OTUs were assigned to the genus Pyramimonas, which include bigger (≈ 5-10 µm) nanoflagellates that can be usually identified by microscopy by the presence of four flagella (Throndsen, 1996). The detection of OTUs assigned to four different cryptophyte genera (i.e., Leucocryptos, Teleaulax, Hanusia, and Katablepharis), in the Argentine Sea is noteworthy, as to our knowledge, these genera had not been previously recorded in this region. This finding suggests a greater diversity of Cryptophytes than previously documented. Despite their relatively large cell size (≈ 10 µm) and their significant contribution to phytoplankton biomass in cold-water regions (Brown et al., 2021), cryptophytes have received limited attention in the Argentine Sea. With the exception of Silva et al. (2009), who mentioned the occurrence of the genera Hemiselmis and Plagioselmis in the northern Argentine Sea, other microscopic studies have typically referred to cryptophytes in this region as “unidentified cryptophytes” (e.g., de Souza et al., 2012; Antacli et al., 2018; Guinder et al., 2018). However, it is important to note that, thus far, no comprehensive examination of cryptophytes using electron microscopy has been conducted in the Argentine Sea.
5 Conclusions
This study provided refined information on biodiversity and environment related variability of phytoplankton communities during austral summer for three different frontal areas in an unprecedented approach for the Argentine Sea. Based on integrated information from microscopic counts, biomass estimation, and 18S sequences phytoplankton community variability could be mainly attributed to the latitudinal gradient along the three frontal areas. To the north, there was a predominance in terms of biomass and taxonomic composition of chlorophytes (small flagellates), dinoflagellates increased at mid latitude, while to the south, diatoms dominated. The use of metabarcoding significantly increased the detection of phytoplankton species richness and our understanding of biodiversity for the Argentine Sea. It allowed the identification of a broad range of dinoflagellates, diatoms, as well as smaller flagellate or coccoid organisms that are beyond the scope of light microscopy, and which have not been described for the Argentine Sea. Some of these smaller organisms, such as the chlorophyte Pycnococcus provasolii, play a crucial role in the southwestern Atlantic ecosystem, particularly in the formation of significant summer blooms. Our study shows the value of combining the strengths of metabarcoding and microscopy to improve our understanding of marine ecosystems, although the weaknesses of the methods should always be considered.
Data availability statement
Raw sequences generated in this study have been deposited at the European Nucleotide Archive (ENA) using the data brokerage service of the German Federation for Biological Data (Diepenbroek et al., 2014) with the accession number PRJEB72201.
Author contributions
GA: Conceptualization, Formal analysis, Funding acquisition, Investigation, Writing – original draft, Writing – review & editing. EF: Formal analysis, Writing – original draft, Writing – review & editing. PS: Formal analysis, Writing – review & editing. MM: Formal analysis, Writing – review & editing. JA: Formal analysis, Writing – review & editing. MF: Writing – review & editing. KM: Formal analysis, Writing – review & editing. FB: Formal analysis, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Council for Scientific and Technical Research [CONICET PIP 1195], the National Agency for Scientific and Technological Promotion [ANPCyT PICT 2719], and FONDECYT 11230767.
Acknowledgments
We thank the support of the crew of the R/V Bernardo Houssay (Prefectura Naval Argentina), Captain Fernando Dos Santos. Furthermore, we thank Kerstin Korte and Swantje Ziemann for excellent technical assistance, and Stefan Neuhaus and Stephan Frickenhaus (Scientific Computing, AWI) are acknowledged for excellent technical support of the sequence analyses. Finally, we want to thank both reviewers for their constructive comments on the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2023.1306336/full#supplementary-material
References
Abad D., Albaina A., Aguirre M., Laza-Martínez A., Uriarte I., Iriarte A., et al. (2016). Is metabarcoding suitable for estuarine plankton monitoring? A comparative study with microscopy. Mar. Biol. 163, 1–13. doi: 10.1007/s00227-016-2920-0
Acha E. M., Mianzan H. W., Guerrero R. A., Favero M., Bava J. (2004). Marine fronts at the continental shelves of austral South America: physical and ecological processes. J. Mar. Syst. 44, 83–105. doi: 10.1016/j.jmarsys.2003.09.005
Almandoz G. O., Fabro E., Ferrario M., Tillmann U., Cembella A., Krock B. (2017). Species occurrence of the potentially toxigenic diatom genus Pseudo-nitzschia and the associated neurotoxin domoic acid in the Argentine Sea. Harmful Algae 63, 45–55. doi: 10.1016/j.hal.2017.01.007
Almandoz G. O., Ferrario M. E., Ferreyra G. A., Schloss I. R., Esteves J. L., Paparazzo F. E. (2007). The genus Pseudo-nitzschia (Bacillariophyceae) in continentalshelf waters of Argentina (Southwestern Atlantic Ocean, 38–55°S). Harmful Algae 6, 93–103. doi: 10.1016/j.hal.2006.07.003
Almandoz G. O., Hernando M. P., Ferreyra G. A., Schloss I. R., Ferrario M. E. (2011). Seasonal phytoplankton dynamics in extreme southern South America (Beagle channel, Argentina). J. Sea Res. 66, 47–57. doi: 10.1016/j.seares.2011.03.005
Andersson A., Zhao L., Brugel S., Figueroa D., Huseby S. (2023). Metabarcoding vs Microscopy: Comparison of methods to monitor phytoplankton communities. ACS ES&T Water 3, 2671–2680. doi: 10.1021/acsestwater.3c00176
Antacli J. C., Silva R. I., Jaureguizar A. J., Hernández D. R., Mendiolar M., Sabatini M. E., et al. (2018). Phytoplankton and protozooplankton on the southern Patagonian shelf (Argentina, 47–55°S) in late summer: Potentially toxic species and community assemblage structure linked to environmental features. J. Sea Res. 140, 63–80. doi: 10.1016/j.seares.2018.07.012
Balch W. M., Drapeau D. T., Bowler B. C., Lyczkowski E. R., Lubelczyk L. C., Painter S. C., et al. (2014). Surface biological, chemical, and optical properties of the Patagonian Shelf coccolithophore bloom, the brightest waters of the Great Calcite Belt. Limnol. Oceanogr. 59, 1715–1732. doi: 10.4319/lo.2014.59.5.1715
Balech E. (1988). Los dinoflagelados del Atlántico sudoccidental. Publ Espec Inst Esp Oceanogr (Madrid: Ministerio de Agricultura, Pesca y Alimentación).
Balech E. (2002). “Dinoflagelados tecados tóxicos del cono sur americano,” in Floraciones Algales Nocivas en el Cono Sur Americano. Eds. Sar E. A., Ferrario M. E., Reguera B. (Vigo: Instituto Español Oceanográfico de Madrid), 125–144.
Bhaud Y., Salmon J. M., Soyer-Gobillard M. O. (1991). The complex cell cycle of the dinoflagellate protoctist Crypthecodinium cohnii as studied in vivo and by cytofluorimetry. J. Cell Sci. 100, 675–682. doi: 10.1242/jcs.100.3.675
Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boutrup P. V., Moestrup Ø., Tillmann U., Daugbjerg N. (2017). Ultrastructure and Phylogeny of Kirithra asteri gen. et sp. nov. (Ceratoperidiniaceae, Dinophyceae)—a Free-Living, Thin-Walled Marine Photosynthetic Dinoflagellate from Argentina. Protist 168, 586–611. doi: 10.1016/j.protis.2017.08.001
Boyer T., Garcia H., Locarnini R., Zweng M., Mishonov A., Reagan J., et al. (2018). World Ocean Atlas 2018 (NOAA National Centers for Environmental Information). Available at: https://www.ncei.noaa.gov/archive/accession/NCEI-WOA18.
Brown M. S., Bowman J. S., Lin Y., Feehan C. J., Moreno C. M., Cassar N., et al. (2021). Low diversity of a key phytoplankton group along the West Antarctic Peninsula. Limnol. Oceanogr. 66, 2470–2480. doi: 10.1002/lno.11765
Burkholder J. M., Glasgow H. B., Deamer-Mella N. (2001). Overview and present status of the toxic Pfiesteria complex (Dinophyceae). Phycologia 40, 186–214. doi: 10.2216/i0031-8884-40-3-186.1
Burki F., Sandin M. M., Jamy M. (2021). Diversity and ecology of protists revealed by metabarcoding. Curr. Biol. 31, R1267–R1280. doi: 10.1016/j.cub.2021.07.066
Cameron E. S., Schmidt P. J., Tremblay B. J. M., Emelko M. B., Müller K. M. (2021). Enhancing diversity analysis by repeatedly rarefying next generation sequencing data describing microbial communities. Sci. Rep. 11 (1), 22302. doi: 10.1038/s41598-021-01636-1
Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carreto J. I., Benavides H. R., Negri R. M., Glorioso P. D. (1986). Toxic red-tide in the Argentine Sea. Phytoplankton distribution and survival of the toxic dinoflagellate Gonyaulax excavata in a frontal area. J. Plankton Res. 8, 15–28. doi: 10.1093/plankt/8.1.15
Carreto J. I., Carignan M. O., Montoya N. G., Cucchi Colleoni A. D. (2007). Ecología del fitoplancton en los sistemas frontales del Mar Argentino. El Mar. Argentino y sus recursos pesqueros 5, 11–31.
Carreto J., Lutz V. A., Carignan M. O., Colleoni A. D., De Marco S. G. (1995). Hydrography and chlorophyll a in a transect from the coast to the shelf-break in the Argentinian Sea. Cont. Shelf Res. 15, 315–336. doi: 10.1016/0278-4343(94)E0001-3
Carreto J. I., Montoya N., Akselman R., Carignan M. O., Silva R. I., Colleoni D. A. (2008). Algal pigment patterns and phytoplankton assemblages in different water masses of the Río de la Plata maritime front. Cont. Shelf Res. 28, 1589–1606. doi: 10.1016/j.csr.2007.02.012
Carreto J. I., Montoya N. G., Carignan M. O., Akselman R., Acha E. M., Derisio C. (2016). Environmental and biological factors controlling the spring phytoplankton bloom at the Patagonian shelf-break front–degraded fucoxanthin pigments and the importance of microzooplankton grazing. Prog. Oceanogr. 146, 1–21. doi: 10.1016/j.pocean.2016.05.002
Cefarelli A. O., Ferrario M. E., Almandoz G. O., Atencio A. G., Akselman R., Vernet M. (2010). Diversity of the diatom genus Fragilariopsis in the Argentine Sea and Antarctic waters: morphology, distribution and abundance. Pol. Biol. 33, 1463–1484. doi: 10.1007/s00300-010-0794-z
Decelle J., Carradec Q., Pochon X., Henry N., Romac S., Mahe F., et al. (2018). Worldwide occurrence and activity of the reef- building coral symbiont Symbiodinium in the open ocean. Curr. Biol. 28, 1–9. doi: 10.1016/j.cub.2018.09.024
de Souza M. S., Mendes C. R. B., Garcia V. M. T., Pollery R., Brotas V. (2012). Phytoplankton community during a coccolithophorid bloom in the Patagonian Shelf: microscopic and high-performance liquid chromatography pigment analyses. J. Mar. Biolog. Assoc. U.K. 92, 13–27. doi: 10.1017/S0025315411000439
De Vargas C., Audic S., Henry N., Decelle J., Mahé F., Logares R., et al. (2015). Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605. doi: 10.1126/science.1261605
Diepenbroek M., Glöckner F., Grobe P., Güntsch A., Huber R., König-Ries B., et al. (2014). “Towards an Integrated Biodiversity and Ecological Research Data Management and Archiving Platform: The German Federation for the Curation of Biological Data (GFBio),” in Informatik 2014 – Big Data Komplexität meistern. GI-Edition: Lecture Notes in Informatics (LNI) – Proceedings. Eds. Plödereder E., Grunske L., Schneider E., Ull D. (Bonn: Köllen Verlag), 1711–1724.
Dodge J. D., Toriumi S. (1993). A taxonomic revision of the Diplopsalis group (Dinophyceae). Bot. Mar. 36, 137–147. doi: 10.1515/botm.1993.36.2.137
Edgar R. C., Haas B. J., Clemente J. C., Quince C., Knight R. (2011). UCHIME improves sensitivity and speed of chi-mera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Fabro E., Almandoz G. O. (2021). Field observations on rare or unnoticed dinoflagellates from the Argentine Sea. Bol. Soc Argent. Bot. 56, 123–140. doi: 10.31055/1851.2372.v56.n2.31421
Ferrario M. E., Almandoz G. O., Cefarelli A. O., Beszteri B., Akselman R., Fabro E., et al. (2018). Shionodiscus gaarderae sp. nov. (Thalassiosirales, Thalassiosiraceae), a bloom-producing diatom from the southwestern Atlantic Ocean, and emendation of Shionodiscus bioculatus var. bioculatus. Diatom Res. 33, 25–37. doi: 10.1080/0269249X.2017.1423112
Ferrario M. E., Sar E. A. (1988). Marine diatoms from Chubut (Argentina Republic) Centrales II-Thalassiosira. Rev. Bras. Biol. 48, 421–429.
Garcia C. A. E., Garcia V. M. T., Dogliotti A. I., Ferreira A., Romero S. I., Mannino A., et al. (2011). Environmental conditions and bio-optical signature of a coccolithophorid bloom in the patagonian shelf. J. Geophys. Res. 116 (C3). doi: 10.1029/2010JC006595
Gloor G. B., Macklaim J. M., Pawlowsky-Glahn V., Egozcue J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8, 2224.
Gómez F., Moreira D., López-García P. (2009). Life cycle and molecular phylogeny of the dinoflagellates Chytriodinium and Dissodinium, ectoparasites of copepod eggs. Eur. J. Protistol. 45, 260–270. doi: 10.1016/j.ejop.2009.05.004
Gonçalves-Araujo R., de Souza M. S., Mendes C. R. B., Tavano V. M., Garcia C. A. (2016). Seasonal change of phytoplankton (spring vs. summer) in the southern Patagonian shelf. Cont. Shelf Res. 124, 142–152. doi: 10.1016/j.csr.2016.03.023
Gran-Stadniczeñko S., Egge E., Hostyeva V., Logares R., Eikrem W., Edvardsen B. (2019). Protist diversity and seasonal dynamics in Skagerrak plankton communities as revealed by metabarcoding and microscopy. J. Eukaryot. Microbiol. 66, 494–513. doi: 10.1111/jeu.12700
Grzebyk D., Audic S., Lasserre B., Abadie E., de Vargas C., Bec B. (2017). Insights into the harmful algal flora in northwestern Mediterranean coastal lagoons revealed by pyrosequencing metabarcodes of the 28S rRNA gene. Harmful Algae 68, 1–16. doi: 10.1016/j.hal.2017.06.003
Guerrero R. A., Baldoni A. G., Benavides H. R. (1999). Oceanographic conditions at the southern end of the Argentine continental slope. Inidep documento científico 5, 7–22.
Guillard R. R., Keller M. D., O’Kelly C. J., Floyd G. L. (1991). Pycnococcus provasolii gen. et sp. nov., a coccoid prasinoxanthin-containing phytoplankter from the western north Atlantic and Gulf of Mexico. J. Phycol. 27, 39–47. doi: 10.1111/j.0022-3646.1991.00039.x
Guillou L., Chrétiennot-Dinet M. J., Medlin L. K., Claustre H., Goër S. L. D., Vaulot D. (1999). Bolidomonas: a new genus with two species belonging to a new algal class, the Bolidophyceae (Heterokonta). J. Phycol. 35, 368–381. doi: 10.1046/j.1529-8817.1999.3520368.x
Guinder V. A., Malits A., Ferronato C., Krock B., Garzón-Cardona J., Martínez A. (2020). Microbial plankton configuration in the epipelagic realm from the Beagle Channel to the Burdwood Bank, a Marine Protected Area in Sub-Antarctic waters. PloS One 15, e0233156. doi: 10.1371/journal.pone.0233156
Guinder V. A., Tillmann U., Krock B., Delgado A. L., Krohn T., Garzon Cardona J. E., et al. (2018). Plankton multiproxy analyses in the Northern Patagonian Shelf, Argentina: Community structure, phycotoxins, and characterization of toxic Alexandrium strains. Front. Mar. Sci. 5, 394. doi: 10.3389/fmars.2018.00394
Guiry M. D. (2022). AlgaeBase. Eds. Guiry M. D., Guiry G. M. (Galway: World-wide electronic publication, National University of Ireland). Available at: https://www.algaebase.org.
Hanžek N., Gligora Udovič M., Kajan K., Borics G., Várbíró G., Stoeck T., et al. (2023). Comparative identification of phytoplankton taxonomic and functional group approach in karst lakes using classical microscopy and eDNA metabarcoding for ecological status assessment. Hydrobiologia, 1–20.
Hasegawa T., Miyashita H., Kawachi M., Ikemoto H., Kurano N., Miyachi S., et al. (1996). Prasinoderma coloniale gen. et sp. nov., a new pelagic coccoid prasinophyte from the western Pacific Ocean. Phycologia 35, 170–176. doi: 10.2216/i0031-8884-35-2-170.1
Hoppenrath M. (2000). Morphology and taxonomy of Sinophysis (Dinophyceae, Dinophysiales) including two new marine sand-dwelling species from the North German Wadden Sea. Eur. J. Phycol. 35, 153–162. doi: 10.1080/09670260010001735741
Kaczmarska I., Mather L., Luddington I. A., Muise F., Ehrman J. M. (2014). Cryptic diversity in a cosmopolitan diatom known as Asterionellopsis glacialis (Fragilariaceae): Implications for ecology, biogeography, and taxonomy. Am. J. Bot. 101, 267–286. doi: 10.3732/ajb.1300306
Kang N. S., Jeong H. J., Moestrup O., Shin W., Nam S. W., Park J. Y., et al. (2010). Description of a newplanktonic mixotrophic dinoflagellate Paragymnodinium shiwhaensen n. gen., n. sp. from the coastal waters off western Korea: morphology, pigments, and ribosomal DNA gene sequence. J. Eukaryot. Microbiol. 57, 121–144. doi: 10.1111/j.1550-7408.2009.00462.x
Lavrinienko A., Jernfors T., Koskimäki J. J., Pirttilä A. M., Watts P. C. (2021). Does intraspecific variation in rDNA copy number affect analysis of microbial communities? Trends Microbiol. 29, 19–27. doi: 10.1016/j.tim.2020.05.019
Le Bescot N., Mahé F., Audic S., Dimier C., Garet M. J., Poulain J., et al. (2016). Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environ. Microbiol. 18, 609–626. doi: 10.1111/1462-2920.13039
Locarnini R. A., Mishonov A. V., Baranova O. K., Boyer T. P., Zweng M. M., Garcia H. E., et al. (2019). World Ocean Atlas 2018, Volume 1: Temperature Vol. 81. Ed. Mishonov A. (NOAA Atlas NESDIS). 52pp.
Mäki A., Salmi P., Mikkonen A., Kremp A., Tiirola M. (2017). Sample preservation, DNA or RNA extraction and data analysis for high-throughput phytoplankton community sequencing. Front. Microbiol. 8, 1848.
Mahé F., Rognes T., Quince C., De Vargas C., Dunthorn M. (2015). Swarm v2: highly-scalable and high-resolution amplicon clustering. PeerJ 3, e1420. doi: 10.7717/peerj.1420
Massana R., Castresana J., Balagué V., Guillou L., Romari K., Groisillier A., et al. (2004). Phylogenetic and ecological analysis of novel marine stramenopiles. Appl. Environ. Microbiol. 70, 3528–3534. doi: 10.1128/AEM.70.6.3528-3534.2004
Metfies K., Hessel J., Klenk R., Petersen W., Wiltshire K. H., Kraberg A. (2020). Uncovering the intricacies of microbial community dynamics at Helgoland Roads at the end of a spring bloom using automated sampling and 18S meta-barcoding. PloS One 15, e0233921. doi: 10.1371/journal.pone.0233921
Moestrup Ø., Lindberg K., Daugbjerg N. (2009). Studies on woloszynskioid dinoflagellates IV: the genus Biecheleria gen. nov. Phycol. Res. 57, 203–220. doi: 10.1111/j.1440-1835.2009.00540.x
Nunes S., Perez G. L., Latasa M., Zamanillo M., Delgado M., Ortega-Retuerta E., et al. (2019). Size fractionation, chemotaxonomic groups and bio-optical properties of phytoplankton along a transect from the Mediterranean Sea to the SW Atlantic Ocean. Sci. Mar. 83, 87–109. doi: 10.3989/scimar.04866.10A
Odebrecht C., Du Preez D. R., Abreu P. C., Campbell E. E. (2013). Surf zone diatoms: A review of the drivers, patterns and role in sandy beaches food chains. Estuar. Coast. Shelf Sci. 150, 24–35. doi: 10.1016/j.ecss.2013.07.011
Oksanen J. (2015). Multivariate analysis of ecological communities in R: vegan tutorial 1–43. doi: 10.1016/0169-5347(88)90124-3
Paparazzo F. E., Bianucci L., Schloss I. R., Almandoz G. O., Solís M., Esteves J. L. (2010). Cross-frontal distribution of inorganic nutrients and chlorophyll-a on the Patagonian Continental Shelf of Argentina during summer and fall. Rev. Biol. Mar. Oceanogr. 45, 107–119. doi: 10.4067/S0718-19572010000100010
Phan-Tan L., Nguyen-Ngoc L., Doan-Nhu H. (2016). Species diversity of sections conica and tabulata in the genus Protoperidinium (Dinophyceae) from tropical waters of the South China Sea. Nova Hedwigia 103, 515–545. doi: 10.1127/nova_hedwigia/2016/0369
Pierella Karlusich J. J., Ibarbalz F. M., Bowler C. (2020). Phytoplankton in the Tara ocean. Ann. Rev. Mar. Sci. 12, 233–265. doi: 10.1146/annurev-marine-010419-010706
Piola A. R. (2008). Oceanografıa fısica. Estado de Conservación del Mar Patagónico y áreas de influencia (Puerto Madryn, Argentina: Edición del Foro), 1–21.
Ramond P., Siano R., Schmitt S., de Vargas C., Marié L., Mémery L., et al. (2021). Phytoplankton taxonomic and functional diversity patterns across a coastal tidal front. Sci. Rep. 11, 1–15. doi: 10.1038/s41598-021-82071-0
Rivas A. L., Dogliotti A. I., Gagliardini D. A. (2006). Seasonal variability in satellite-measured surface chlorophyll in the Patagonian Shelf. Cont. Shelf Res. 26, 703–720. doi: 10.1016/j.csr.2006.01.013
Ruggiero M. V., Sarno D., Barra L., Kooistra W. H., Montresor M., Zingone A. (2015). Diversity and temporal pattern of Pseudo-nitzschia species (Bacillariophyceae) through the molecular lens. Harmful Algae 42, 15–24.
Santi I., Kasapidis P., Karakassis I., Pitta P. (2021). A comparison of DNA metabarcoding and microscopy methodologies for the study of aquatic microbial eukaryotes. Diversity 13, 180. doi: 10.3390/d13050180
Santoferrara L. F. (2019). Current practice in plankton metabarcoding: optimization and error management. J. Plankton Res. 41, 571–582. doi: 10.1093/plankt/fbz041
Sar E. A., Sterrenburg F. A., Sunesen I. (2014). Pleurosigma hinzianum sp. nov. and P. frenguellianum sp. nov. (Pleurosigmataceae, Bacillariophyta) from Argentinean coastal waters, in comparison with P. amara Stidolph and P. elongatum W. Smith. Eur. J. Phycol. 49, 151–164. doi: 10.1080/09670262.2014.898101
Sar E. A., Sunesen I., Lavigne A. S. (2002). The diatom genus Thalassiosira: species from the northern San Matías Gulf (Río Negro, Argentina). Nova Hedwigia 74, 373–386. doi: 10.1127/0029-5035/2002/0074-0373
Seenivasan R., Sausen N., Medlin L. K., Melkonian M. (2013). Picomonas judraskeda gen. et sp. nov.: the first identified member of the Picozoa phylum nov., a widespread group of picoeukaryotes, formerly known as ‘picobiliphytes’. PloS One 8, e59565. doi: 10.1371/journal.pone.0059565
Shalchian-Tabrizi K., Kauserud H., Massana R., Klaveness D., Jakobsen K. S. (2007). Analysis of environmental 18S ribosomal RNA sequences reveals unknown diversity of the cosmopolitan phylum Telonemia. Protist 158, 173–180. doi: 10.1016/j.protis.2006.10.003
Siano R., Montresor M., Probert I., Not F., de Vargas C. (2010). Pelagodinium gen. nov. and P. béii comb. nov., a dinoflagellate symbiont of planktonic foraminifera. Protist 161, 385–399. doi: 10.1016/j.protis.2010.01.002
Silva R., Negri R., Lutz V. (2009). Summer succession of ultraphytoplankton at the EPEA coastal station (Northern Argentina). J. Plankton Res. 31, 447–458. doi: 10.1093/plankt/fbn128
Sunesen I., Hernández Becerril D. U., Sar E. A. (2008). Marine diatoms from Buenos Aires coastal waters (Argentina). V. Species of the genus Chaetoceros. Rev. Biol. Mar. Oceanogr. 43, 303–326. doi: 10.4067/S0718-19572008000200009
Sunesen I., Rodríguez F., Tardivo Kubis J. A., Aguiar Juárez D., Risso A., Lavigne A. S., et al. (2020). Morphological and molecular characterization of Heterocapsa claromecoensis sp. nov. (Peridiniales, Dinophyceae) from Buenos Aires coastal waters (Argentina). Eur. J. Phycol. 55, 490–506. doi: 10.1080/09670262.2020.1750059
Takano Y., Horiguchi T. (2004). Surface ultrastructure and molecular phylogenetics of four unarmored heterotrophic dinoflagellates, including the type species of the genus Gyrodinium (Dinophyceae). Phycol. Res. 52, 107–116. doi: 10.1111/j.1440-1835.2004.tb00319.x
Throndsen J. (1996). “The planktonic marine flagellates,” in Identifying marine phytoplankton. Ed. Tomas C. R. (San Diego: Academic Press), 591–730.
Tillmann U., Gottschling M., Guinder V., Krock B. (2018). Amphidoma parvula (Amphidomataceae), a new planktonic dinophyte from the Argentine Sea. Eur. J. Phycol. 53, 14–28. doi: 10.1080/09670262.2017.1346205
Turk Dermastia T., Vascotto I., Francé J., Stanković D., Mozetič P. (2023). Evaluation of the rbcL marker for metabarcoding of marine diatoms and inference of population structure of selected genera. Front. Microbiol. 14, 1071379.
Utermöhl H. (1958). Zur vervollkommnung der quantitativen phytoplankton-methodik. Mitt. Int. Ver. Theor. Angew. Limnol. 9, 1–38.
Uyua N. M., Manrique J. M., Jones L. R. (2014). An optimized DNA extraction protocol for benthic Didymosphemia geminata. J. Microbiol. Methods 104, 12–18. doi: 10.1016/j.mimet.2014.06.007
Yuasa T., Horiguchi T., Mayama S., Takahashi O. (2016). Gymnoxanthella radiolariae gen. et sp. nov. (Dinophyceae), a dinoflagellate sym-biont from solitary polycystine radiolarians. J. Phycol. 52, 89–104. doi: 10.1111/jpy.12371
Zingone A., Sarno D., Siano R., Marino D. (2011). The importance and distinctiveness of small-sized phytoplankton in the Magellan Straits. Pol. Biol. 34, 1269–1284. doi: 10.1007/s00300-010-0937-2
Keywords: metabarcoding, dinoflagelates, unidentified Gyrodinium, chlorophyte blooms, Pycnococcus provasolii
Citation: Almandoz GO, Fabro E, Sprong P, Mascioni M, Antoni J, Ferrario M, Metfies K and Barrera FM (2024) Metabarcoding and microscopy characterization of phytoplankton from frontal areas of the Argentine Sea. Front. Mar. Sci. 10:1306336. doi: 10.3389/fmars.2023.1306336
Received: 03 October 2023; Accepted: 04 December 2023;
Published: 07 February 2024.
Edited by:
David Stanković, National Institute of Biology, SloveniaReviewed by:
Timotej Turk Dermastia, National Institute of Biology, SloveniaStacy Louise Deppeler, National Institute of Water and Atmospheric Research (NIWA), New Zealand
Copyright © 2024 Almandoz, Fabro, Sprong, Mascioni, Antoni, Ferrario, Metfies and Barrera. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gastón Osvaldo Almandoz, galmandoz@fcnym.unlp.edu.ar; Facundo Matias Barrera, facundobarrera182@gmail.com
 Gastón Osvaldo Almandoz
Gastón Osvaldo Almandoz Elena Fabro1,2
Elena Fabro1,2  Martina Mascioni
Martina Mascioni Katja Metfies
Katja Metfies Facundo Matias Barrera
Facundo Matias Barrera