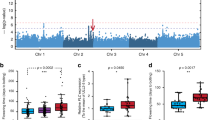
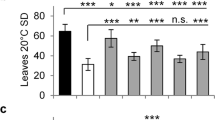
Abstract
Variation of flowering time is found in the natural populations of many plant species. The underlying genetic variation, mostly of a quantitative nature, is presumed to reflect adaptations to different environments contributing to reproductive success. Analysis of natural variation for flowering time in Arabidopsis thaliana has identified several quantitative trait loci (QTL)1, which have yet to be characterized at the molecular level. A major environmental factor that determines flowering time is photoperiod or day length, the length of the light period, which changes across the year differently with geographical latitude2. We identified the EDI locus as a QTL partly accounting for the difference in flowering response to the photoperiod between two Arabidopsis accessions: the laboratory strain Landsberg erecta (Ler), originating in Northern Europe, and Cvi, collected in the tropical Cape Verde Islands3. Positional cloning of the EDI QTL showed it to be a novel allele of CRY2, encoding the blue-light photoreceptor cryptochrome-2 that has previously been shown to promote flowering in long-day (LD) photoperiods4. We show that the unique EDI flowering phenotype results from a single amino-acid substitution that reduces the light-induced downregulation of CRY2 in plants grown under short photoperiods, leading to early flowering.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Koornneef, M., Alonso-Blanco, C., Peeters, A.J.M. & Soppe, W. Genetic control of flowering time in Arabidopsis. Ann. Rev. Plant Physiol. Plant Mol. Biol. 49, 345–370 (1998).
Thomas, B. & Vince-Prue, D. Photoperiodism in Plants (Academic Press, New York, 1997).
Alonso-Blanco, C., El-Assal, S.E-D., Coupland, G. & Koornneef, M. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Island ecotypes of Arabidopsis thaliana. Genetics 149, 749–764 (1998).
Guo, H., Yang, H., Mockler, T.C. & Lin, C. Regulation of flowering time by Arabidopsis photoreceptors. Science 279, 1360–1363 (1998).
Karlsson, B.H., Sills, G.R. & Nienhuis, J. Effect of photoperiod and vernalization on the number of leaves at flowering in 32 Arabidopsis thaliana (Brassicaceae) ecotypes. Am. J. Bot. 80, 646–648 (1993).
Koornneef, M., Hanhart, C.J. & van der Veen, J.H. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 229, 57–66 (1991).
Jansen, R.C., Van Ooijen, J.W., Stam, P., Lister, C. & Dean, C. Genotype by environment interaction in genetic mapping of multiple quantitative trait loci. Theor. Appl. Genet. 91, 33–37 (1995).
Neff, M.M., Neff, J.D., Chory, J. & Pepper, E. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 14, 387–392 (1998).
Lin, C. et al. Enhancement of blue-light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl Acad. Sci. USA 95, 2686–2690 (1998).
Ahmad, M., Jarillo, J.A. & Cashmore, A.R. Chimeric protein between cry1 and cry2 Arabidopsis blue light photoreceptors indicate overlapping functions and varying protein stability. Plant Cell 10, 197–207 (1998).
Bernier, G., Havelange, A., Houssa, C., Petitjean, A. & Lejeune, P. Physiological signals that induce flowering. Plant Cell 5, 1147–1155 (1993).
Bradley, D., Ratcliffe, O., Vincent, C., Carpenter, R. & Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science 275, 80–83 (1997).
Mockler, T.C., Guo, H., Yang, H., Duong, H. & Lin, C. Antagonistic action of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126, 2073–2082 (1999).
Mizoguchi, T. & Coupland, G. ZEITLUPE and FKF1: novel connections between flowering time and circadian clock control. Trends Plant Sci. 5, 409–411 (2000).
Devlin, P.F. & Kay, S.A. Cryptochromes are required for phytochrome signalling to the circadian clock but not for rhythmicity. Plant Cell 12, 2499–2509 (2000).
Swarup, K. et al. Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J. 20, 67–77 (1999).
Suarez-Lopez, P. et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410, 1116–1120 (2001).
Johanson, U. et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347 (2000).
Michaels, S.D. & Amasino, R.M. Flowering Locus C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Yano, M. et al. Hd1, a major photoperiod senstivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2483 (2000).
Takahashi, Y., Shomura, A., Sasaki, T. & Yano, M. Hd6, a rice quantitative trait locus involved in photoperiod sensitivity, encodes the α subunit of protein kinase CK2. Proc. Natl Acad. Sci. USA 98, 7922–7927 (2001).
Wang, R.L., Stec, A., Hey, J., Lukens, L. & Doebley, J. The limits of selection during maize domestication. Nature 398, 236–239 (1999).
Frary, A. et al. Fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289, 85–88 (2000).
Fridman, E., Pleban, T. & Zamir, D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl Acad. Sci. USA 97, 4718–4723 (2000).
Kliebenstein, D.J., Lambrix, V.M., Reichelt, M., Gershenzon, J. & Mitchell-Olds, T. Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13, 681–693 (2001).
Maloof, J.N. et al. Natural variation in light sensitivity of Arabidopsis. Nature Genet. 29, 441–446 (2001).
Koornneef, M., Hanhart, C., van Loenen-Martinet, P. & Blankestijn-de Vries, H. The effect of daylength on the transition to flowering in phytochrome-deficient, late-flowering and double mutants of Arabidopsis thaliana. Physiol. Planta 95, 260–266 (1995).
Van Tuinen, A., Kerckhoffs, L.H.J., Nagatani, A., Kendrick, R.E. & Koornneef, M. Far-red light-insensitive, phytochrome A–deficient mutants of tomato. Mol. Gen. Genet. 246, 133–141 (1995).
Peters, J.L., Schreuder, M.E.L., Verduin, S.J.W. & Kendrick, R.E. Physiological characterization of a high-pigment mutant of tomato. Photochem. Photobiol. 56, 75–82 (1992).
Bechtold, N., Ellis, J. & Pelletier, G. In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. 316, 1194–1199 (1993).
Lazo, G.R., Stein, P.A. & Ludwig, R.A. A DNA transformation–competent Arabidopsis genomic library in Agrobacterium. Biotechnology 9, 963–967 (1991).
Chou, I.T. & Gasser, C.S. Characterization of the cyclophilin gene family of Arabidopsis thaliana and phylogenetic analysis of known cyclophilin protein. Plant Mol. Biol. 35, 873–892 (1997).
Guan, K. & Dixon, J.G. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion protein with glutathione S-transferase. Anal. Biochem. 192, 262–267 (1991).
Sessa, G., Yang, C.-X., Raz, V., Eyal, Y. & Fluhr, R. Dark induction and subcellular localization of the pathogenesis related PRB-1b protein. Plant Mol. Biol. 28, 537–547 (1995).
Raz, V. & Ecker, J.R. Regulation of differential growth in the apical hook of Arabidopsis. Development 126, 3661–3668 (1999).
Cashmore, A.R., Jarillo, J.A., Wu, Y.J. & Liu, D. Cryptochrome: blue light receptors for plants and animals. Science 284, 760–765 (1999).
Acknowledgements
We wish to thank A. Miller for providing transgenic seeds expressing the CAB:LUC constructs, J. Jarillo for providing CRY2 cDNA, W. Soppe for his continuous help and support, J. Weller for his assistance in the blue-light experiments and for critical reading of the manuscript, H.H. Offenberg and S. van der Krol for their assistance in developing anti-CRY2 and in the luciferase measurements, respectively, and M. Schreuder and C. Hanhart for help with the light cabinets and greenhouse work. This work was supported by a predoctoral fellowship from the government of Egypt to S.E.-D.E.-A.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
El-Din El-Assal, S., Alonso-Blanco, C., Peeters, A. et al. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat Genet 29, 435–440 (2001). https://doi.org/10.1038/ng767
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng767
This article is cited by
-
Common evolutionary trajectory of short life-cycle in Brassicaceae ruderal weeds
Nature Communications (2023)
-
Parallel reduction in flowering time from de novo mutations enable evolutionary rescue in colonizing lineages
Nature Communications (2022)
-
Environment-mediated mutagenetic interference on genetic stabilization and circadian rhythm in plants
Cellular and Molecular Life Sciences (2022)
-
Characterization of Cry2 genes (CRY2a and CRY2b) of B. napus and comparative analysis of BnCRY1 and BnCRY2a in regulating seedling photomorphogenesis
Plant Molecular Biology (2022)
-
Structural insights into BIC-mediated inactivation of Arabidopsis cryptochrome 2
Nature Structural & Molecular Biology (2020)