Abstract
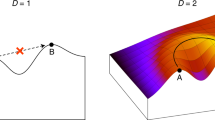
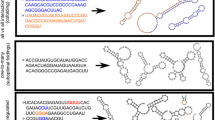
RNA secondary-structure folding algorithms predict the existence of connected networks of RNA sequences with identical secondary structures. Fitness landscapes that are based on the mapping between RNA sequence and RNA secondary structure hence have many neutral paths. A neutral walk on these fitness landscapes gives access to a virtually unlimited number of secondary structures that are a single point mutation from the neutral path. This shows that neutral evolution explores phenotype space and can play a role in adaptation.
Similar content being viewed by others
References
Ekland EH, Szostak JW, Bartel DP (1995) Structurally complex and highly active RNA ligases derived from random RNA sequences. Science 269:364–370
Fontana W, Konings DAM, Stadler PF, Schuster P (1993) Statistics of RNA secondary structures. Biopolymers 33:1389–1404
Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster PA (a) Vienna RNA Package. pub/RNA/ViennaRNA-1.03 ftp.itc.univie.ac.at (Public Domain Software)
Huynen MA, Konings DAM, Hogeweg P (1993) Multiple coding and the evolutionary properties of RNA secondary structures. J Theor Biol 165:251–267
Huynen MA, Stadler PF, Fontana W (1996a) Smoothness within ruggedness: the role of neutrality in adaptation. Proc Natl Acad Sci USA 93:397–401
Huynen MA, Perelson A, Vieira W, Stadler PF (1996b) Base pairing probabilities in a complete HIV-1 genome. J Comp Biol 3:253–274
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Konings DAM, Hogeweg P (1989) Pattern analysis of RNA secondary structures, similarity and consensus of minimal-energy folding. J Mol Biol 207:597–614
Maynard-Smith J (1970) Natural selection and the concept of a protein space. Nature 255:563–564
Provine WB (1986) Sewall Wright and evolutionary biology. University of Chicago Press, Chicago
Schuster P, Fontana W, Stadler PF, Hofacker IL (1994) From sequences to shapes and back: a case study in RNA secondary structures. Proc R Soc Lond (Biol) 255:279–284
Tacker M, Stadler PF, Bornberg-Bauer E, Hofacker IL, Schuster P (1996) Robust properties of RNA secondary structure folding algorithms. Working Paper 96-04-016, Santa Fe Institute
Waterman MS (1978) Secondary structure of single-stranded nucleic acids. Studies on foundations and combinatories. Adv Math Suppl Studies 1:167–212
Wright S (1932) The roles of mutation, inbreeding, crossbreeding and selection in evolution. In: Jones DF (ed) Proceedings of the sixth international congress on genetics, vol 1. pp 356–366, Brooklyn Botanic Garden, New York
Zuker M, Stiegler P (1981) Optimal computer folding of larger RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res 9:133–148
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Huynen, M.A. Exploring phenotype space through neutral evolution. J Mol Evol 43, 165–169 (1996). https://doi.org/10.1007/BF02338823
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02338823