Summary
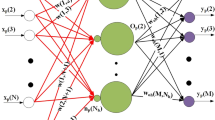
Four different artificial neural network architectures have been tested for their suitability to extract and predict sequence features. For optimization of the network weights an evolutionary computing method has been applied. The networks have feedforward architecture and provide adaptive neural filter systems for pattern recognition in primary structures and sequence classification. The recognition and prediction of signal peptidase cleavage sites ofE. coli periplasmic protein precursors serves as an example for filter development. The primary structures are represented by seven physicochemical residue properties. This amino acid description provides the feature space for network optimization. The properties hydrophobicity, hydrophilicity, side-chain volume, and polarity allowed an accurate classification of the data. A three-layer network architecture reached a learning success of 100%; the highest prediction accuracy in an independent test set of sequences was 97%. This network architecture appears to be most suited for the analysis ofE. coli signal peptidase cleavage sites. Further suggestions about the design and future applications of artificial neural networks for protein sequence analysis are made.
Similar content being viewed by others
References
Bohr H, Bohr J, Brunak S, Cotterill RMJ, Lautrup B, Norskov L, Olsen O, Petersen SB (1988) Protein secondary structure and homology by neural networks. FEBS Lett 241:223–228
Bohr H, Bohr S, Brunak S, Cotterill RMJ, Fredholm H, Lautrup B, Petersen SB (1990) A novel approach to prediction of the 3-dimensional structures of protein backbones byneural networks. FEBS Lett 261:43–46
Cybenko G (1989) Approximation by superpositions of a sigmoidal function. Mathematics of Control, Signals, and Systems 2:303–314
Chothia C (1975) The nature of accessible and buried surfaces in proteins. J Mol Biol 105:1–14
Engelman DA, Steitz TA, Goldman A (1986) Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem 15:321–353
Fasman G (ed) (1989) Prediction of protein structure and the principles of protein conformation. Plenum Press, New York
Gascuel O, Danchin A (1986) Protein export in prokaryotes and eukaryotes: indications of a difference in the mechanism of exportation. J Mol Evol 24:130–142
Hirst JD, Sternberg MJE (1992) Prediction of structural and functional features of protein and nucleic acid sequences by artificial neural networks. Biochemistry 31:7211–7218
Holley LH, Karplus M (1989) Protein secondary structure prediction with a neural network. Proc Natl Acad Sci 86:152–156
Hopp TP, Woods KR (1981) Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA 78:3824–3828
Hornik K, Stinchcombe M, White H (1989) Multilayer feedforward networks are universal approximators. Neural Networks 2:359–366
IntelliGenetics Inc, 700 East Camino Real, Mountain View, CA 94040, USA
Jáhnig F (1990) Structure predictions of membrane proteins are not that bad. Trends Biochem Sci 15:93–95
Jones DD (1975) Amino acid properties and side chain orientation in proteins: A cross correlation approach. J Theor Biol 50:167–183
King RD, Sternberg MJE (1990) Protein secondary structure prediction: a machine learning approach. J Mol Biol 216:441–457
Kosko B (1992) Neural networks and fuzzy systems. Prentice-Hall International, London
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Laforet GA, Kendall DA (1991) Functional limits of conformation, hydrophobicity, and steric constraints in prokaryotic signal peptide cleavage regions. J Biol Chem 266:1326–1334
Levin JM, Gamier J, Biou V, Gibrat JF, Robson B (1988) Secondary structure prediction: combination of three different methods. Protein Engineering 2:185–191
Lohmann R (1992) Structure evolution in neural systems. In: (Soucek B, the Iris Group (eds) Dynamic, genetic and chaotic programming. Wiley & Sons, New York
McInerny JM, Haines KG, Biafore S, Hecht-Nielsen R (1989) Back propagation error surfaces can have local minima. In: International Joint Conference on Neural Networks (Washington 1989), Vol II. IEEE, New York, p 627
Minsky M, Papert S (1988) Perceptrons. MIT Press, Cambridge, MA
Perlman D, Halvorson HA (1983) A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol 167:391–409
Qian N, Sejnowski TJ (1988) Predicting the secondary structure of globlar proteins using neural network models. J Mol Biol 202:865–884
Rechenberg I (1973) Evolutionsstrategie-Optimierung technischer Systeme nach Prinzipien der biologischen Evolution. Frommann-Holzboog, Stuttgart
Rosenblatt F (1962) Principles of neurodynamics. Spartan, New York
Schneider G, Wrede P (1993) Analysis of protein targeting sequence features. Protein Seq Data Anal (in press)
Schulz GE, Schirmer RH (1979) Principles of protein structure. Springer-Verlag, Heidelberg
Stolorz P, Lapedes A, Xia Y (1992) Predicting protein secondary structure using neural net and statistical methods. J Mol Biol 225:363–377
von Heijne G (1983) Patterns of amino acids near signal sequence cleavage sites. Eur J Biochem 133:17–21
von Heijne G (1986) A new method for predicting signal sequence cleavage sites. Nucleic Acids Res 14:4683–4690
Zamyatnin AA (1972) Protein volume in solution. Prog Biophys Mol Biol 24:107–123
Zhang X, Mesirov JP, Waltz DL (1992) Hybrid system for protein secondary structure prediction. J Mol Biol 225:1049–1063
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schneider, G., Wrede, P. Development of artificial neural filters for pattern recognition in protein sequences. J Mol Evol 36, 586–595 (1993). https://doi.org/10.1007/BF00556363
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF00556363