Abstract
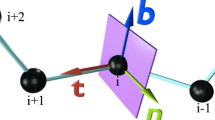
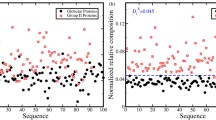
The conformations of protein loops from a non-redundant set of 347 proteins with less than 25% sequence homology have been studied in order to clarify the topological variation of protein loops. Loops have been classified in five types (α-α, α-β, β-α, β-links and β-hairpins) depending on the secondary structures that they embrace. Four variables have been used to describe the loop geometry (3 angles and the end-to-end distance between the secondary structures embracing the loop). Loops with well defined geometry are identified by means of the internal dependency between the geometrical variables by application of information-entropy theory. From this it has been deduced that loops formed by less than 10 residues show an intrinsic dependency on the geometric variables that defines the motif shape. In this interval the most stable loops are found for short connections owing to the entropic energy analysed.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 27 July 1998 / Accepted: 18 September 1998 / Published: 24 November 1998
Rights and permissions
About this article
Cite this article
Martí-Renom, M., Mas, J., Aloy, P. et al. Statistical Analysis of the Loop-Geometry on a Non-Redundant Database of Proteins. J Mol Med 4, 347–354 (1998). https://doi.org/10.1007/s008940050093
Issue Date:
DOI: https://doi.org/10.1007/s008940050093