Abstract
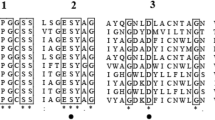
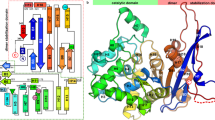
The crystal structure of a pepstatin-insensitive carboxyl proteinase from Pseudomonas sp. 101 (PSCP) has been solved by single-wavelength anomalous diffraction using the absorption peak of bromide anions. Structures of the uninhibited enzyme and of complexes with an inhibitor that was either covalently or noncovalently bound were refined at 1.0–1.4 Å resolution. The structure of PSCP comprises a single compact domain with a diameter of ∼55 Å, consisting of a seven-stranded parallel β-sheet flanked on both sides by a number of helices. The fold of PSCP is a superset of the subtilisin fold, and the covalently bound inhibitor is linked to the enzyme through a serine residue. Thus, the structure of PSCP defines a novel family of serine-carboxyl proteinases (defined as MEROPS S53) with a unique catalytic triad consisting of Glu 80, Asp 84 and Ser 287.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Oda, K., Sugitani, M., Fukuhara, K. & Murao, S. Biochim. Biophys. Acta 923, 463–469 (1987).
Oda, K., Takahashi, T., Tokuda, Y., Shibano, Y. & Takahashi, S. J. Biol. Chem. 269, 26518–26524 (1994).
Ito, M., Narutaki, S., Uchida, K. & Oda, K. J. Biochem. (Tokyo), 125, 210–216 (1999).
Oyama, H., Abe, S., Ushiyama, S., Takahashi, S. & Oda, K. J. Biol. Chem. 274, 27815–27822 (1999).
Davies, D.R. Annu. Rev. Biophys. Biophys. Chem. 19, 189–215 (1990).
Barrett, A.J., Rawlings, N.D. & Woessner, J.F. Handbook of proteolytic enzymes (Academic Press, London; 1998).
Murao, S. et al. J. Biol. Chem. 268, 349–355 (1993).
Shibata, M., Dunn, B.M. & Oda, K. J. Biochem. (Tokyo) 124, 642–647 (1998).
Sleat, D.E. et al. Science 277, 1802–1805 (1997).
Rawlings, N.D. & Barrett, A.J. Biochim. Biophys. Acta 1429, 496–500 (1999).
Lin, L., Sohar, I., Lackland, H. & Lobel, P. J. Biol. Chem. 276, 2249–2255 (2001).
Kwon, H.K., Kim, H. & Ahn, T.I. Korean J. Biol. Sci. 3, 221–228 (1999).
Wright, C.S., Alden, R.A. & Kraut, J. Nature 221, 235–242 (1969).
Holm, L. & Sander, C. J. Mol. Biol. 233, 123–138 (1993).
Kuhn, P. et al. Biochemistry 37, 13446–13452 (1998).
Robertus, J.D., Kraut, J., Alden, R.A. & Birktoft, J.J. Biochemistry 11, 4293–4303 (1972).
Miller, M., Rao, J.K.M., Wlodawer, A. & Gribskov, M.R. FEBS Lett. 328, 275–279 (1993).
Dodson, G. & Wlodawer, A. Trends Biochem. Sci. 23, 347–352 (1998).
Delbaere, L.T. & Brayer, G.D. J. Mol. Biol. 183, 89–103 (1985).
Bullock, T.L., Breddam, K. & Remington, S.J. J. Mol. Biol. 255, 714–725 (1996).
Schechter, I. & Berger, A. Biochem. Biophys. Res. Commun. 27, 157–162 (1967).
Bode, W., Papamokos, E. & Musil, D. Eur. J. Biochem. 166, 673–692 (1987).
Oda, K., Nakatani, H. & Dunn, B.M. Biochim. Biophys. Acta 1120, 208–214 (1992).
Dauter, Z., Li, M. & Wlodawer, A. Acta Crystallogr. D 57, 239–249 (2001).
Oda, K., Fukuda, Y., Murao, S., Uchida, K. & Kainosho, M. Agric. Biol. Chem. 53, 405–415 (2000).
Otwinowski, Z. & Minor, W. Methods Enzymol. 276, 307–326 (1997).
Sheldrick, G.M. In Direct methods for solving macromolecular structures. (ed. Fortier, S.) 401–411 (Kluwer Academic Publishers, Dordrecht; 1998).
de la Fortelle, E. & Bricogne, G. Methods Enzymol. 276, 472–494 (1997).
Cowtan, K.D. & Main, P. Acta Crystallogr. D 52, 43–48 (1996).
Perrakis, A., Morris, R. & Lamzin, V.S. Nature Struct. Biol. 6, 458–463 (1999).
Jones, T.A. & Kieldgaard, M. Methods Enzymol. 277, 173–208 (1997).
Sheldrick, G.M. & Schneider, T.R. Methods Enzymol. 277, 319–343 (1997).
Engh, R. & Huber, R. Acta Crystallogr. A 47, 392–400 (1991).
Kraulis, P.J. J. Appl. Crystallogr. 24, 946–950 (1991).
Carson, M. J. Appl. Crystallogr. 24, 958–961 (1991).
Acknowledgements
We thank A. Barrett for helpful discussions, A. Arthur for editorial comments and J. Alexandratos for help in preparation of the figures. This work was supported in part by a Grant-in-Aid for Scientific Research and a Grant-in-Aid for International Scientific Research (Joint Research) from the Ministry of Education, Science, Sports and Culture of Japan to K.O., by NIH grants to B.M.D., and in part with Federal funds from the National Cancer Institute, National Institutes of Health. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the U. S. Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wlodawer, A., Li, M., Dauter, Z. et al. Carboxyl proteinase from Pseudomonas defines a novel family of subtilisin-like enzymes. Nat Struct Mol Biol 8, 442–446 (2001). https://doi.org/10.1038/87610
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/87610
This article is cited by
-
Crystal structure of a subtilisin-like autotransporter passenger domain reveals insights into its cytotoxic function
Nature Communications (2023)
-
Computational studies of polyurethanases from Pseudomonas
Journal of Molecular Modeling (2021)
-
Structure-function analysis of Sedolisins: evolution of tripeptidyl peptidase and endopeptidase subfamilies in fungi
BMC Bioinformatics (2018)
-
Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses
Neurogenetics (2005)