Abstract
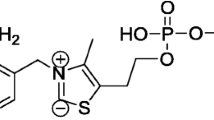
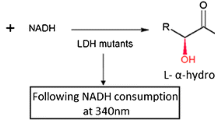
This method gives a general ideal how to use crystallographic information of enzymes to understand reactions catalyzed by these biocatalysts, commonly used by biochemists to produce chiral products. The interactions of three acetoacetic esters with the enzymes L-lactate dehydrogenase and alcohol dehydrogenase were studied through molecular modelling computer program. These artificial substrates have been widely used to produce chiral synthons. Through this methodology it was possible to understand the conformational specificity of these enzymes with respect to the products and how these enzymes can be inhibited by modifying the structures of the artificial substrates. Also, it was possible to predict whether some type of artificial substrate will suffer reduction by cells that contain these dehydrogenases and what kind of configuration (R or S) the final product will have.
Similar content being viewed by others
References
Weijers CAGM, Litjens MJJ, de Bont JAM: Synthesis of optically pure 1,2-epoxypropane by microbial asymmetric reduction of chloro acetone. Appl Microbiol Biotechnol 38: 297–300, 1992
Pereira RS: Baker's yeast: Some biochemical aspects and their influence in biotransformations. Appl Biochem Biotechnol 55: 123–132, 1995
Pereira RS, Parizotto NA, Baranauskas V: Observation of baker's yeast strains used in biotransformation by atomic force microscopy. Appl Biochem Biotechnol 59: 135–143, 1996
Pereira RS: Fermento biológico de padaria (S. ticerevisiae) e Seu Uso em Sínteses Assimétricas. Quím Nova 18: 452–459, 1995
Nakamura K, Kawai Y, Nakajima N, Ohno A: Stereochemical control of microbial reduction 17. A method for controlling the enantioselectivity of reductions with baker's yeast. J Org Chem 56: 4778–4383, 1991
Peters J, Zelinski M, Kula M: Studies on the distribution and regulation of microbial keto ester reductases. Appl Microbiol Biotechnol 38: 334–340, 1992
Russell I, Jones R, Stewart G: In: SK Harlander, TP Labuza (eds). Biotechnology in Food Processing. Noyes Publications, Park Ridge, N.J., USA, 1986, pp 172–185
Shieh W, Gopalan AS, Sih CJ: Stereochemical control in yeast reductions. 5. Characterization of the oxidoreductases involved in the reduction of β-keto esters. J Am Chem Soc 107: 2993–2994, 1985
Zhou BN, Gopalan AS, VanMiddlesworth F, Shieh WR, Sih CJ: Stereochemical control of yeast reductions. 1. Asymmetric synthesis of L-carnitine. J Am Chem Soc 105: 5925–5926, 1983
Jones TA. TOM: In: D Savre (ed). A Graphics Fitting Program for Macromolecules in Computational Crystallography. Clarendon Press, Oxford, 1982, pp 303–317.
Cunningham EB (ed). Mechanisms of Metabolism. Mc-Graw-Hill, New York, USA, 1978, p 526
Voet D, Voet JG: Biochemistry. John Wiley and Sons, New York, USA, 1990, p 445
Carson M: Ribbon models of macromolecules. J Mol Graphics 5: 103–106, 1987
Carson M, Bugg CE: Algorithm for ribbon models of proteins. J Mol Graphics 4: 121–122, 1986
Mackworth JF: The inhibition of thiol enzymes by lachrymators. Biochem J 42: 82–90, 1948
Sorrilha AEPM, Marques M, Joekes I, Morán PJS, Rodrigues JAR: Reduction of phenylketones by immobilized baker's yeast. Bioorg Med Chem Lett 2: 191–196, 1992
Stinson SC: Chiral drugs. Chem Eng News 70: 46–79, 1992
Mori K: Synthesis of optically active pheromones. Tetrahedron 45: 3233–3298, 1989
Hensel R, Mayr U, Yang C: The complete primary structure of the allosteric L-lactate dehydrogenase from L. casei. Eur J Biochem 134: 503–511, 1983
Jörnvall H, Eklund H, Brändén C: Subunit conformation of yeast alcohol dehydrogenase. J Biol Chem 253: 8414–8419, 1978
Eklund H, Brändén C: Structural Comparisons of mammalian, yeast and bacillar alcohol dehydrogenases. J Mol Biol 102: 61–73, 1976
Morrison RT, Boyd RN: Organic Chemistry, 3rd Edition. Allyn and Bacon, Inc., Boston, USA, 1973, pp 124–132
Sicsic S: Enzymes et chimie fine. La Recherche 18: 626–630, 1987
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
de Souza Pereira, R., Pavão, F. & Oliva, G. The use of molecular modelling in the understanding of configurational specificity (R or S) in asymmetric reactions catalyzed by Saccharomyces cerevisiae or isolated dehydrogenases. Mol Cell Biochem 178, 27–31 (1998). https://doi.org/10.1023/A:1006831902849
Issue Date:
DOI: https://doi.org/10.1023/A:1006831902849