Abstract
Autophagy is an evolutionarily conserved catabolic process that recycles nutrients upon starvation and maintains cellular energy homeostasis1,2,3. Its acute regulation by nutrient-sensing signalling pathways is well described, but its longer-term transcriptional regulation is not. The nuclear receptors peroxisome proliferator-activated receptor-α (PPARα) and farnesoid X receptor (FXR) are activated in the fasted and fed liver, respectively4,5. Here we show that both PPARα and FXR regulate hepatic autophagy in mice. Pharmacological activation of PPARα reverses the normal suppression of autophagy in the fed state, inducing autophagic lipid degradation, or lipophagy. This response is lost in PPARα knockout (Ppara−/−, also known as Nr1c1−/−) mice, which are partially defective in the induction of autophagy by fasting. Pharmacological activation of the bile acid receptor FXR strongly suppresses the induction of autophagy in the fasting state, and this response is absent in FXR knockout (Fxr−/−, also known as Nr1h4−/−) mice, which show a partial defect in suppression of hepatic autophagy in the fed state. PPARα and FXR compete for binding to shared sites in autophagic gene promoters, with opposite transcriptional outputs. These results reveal complementary, interlocking mechanisms for regulation of autophagy by nutrient status.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Mizushima, N., Levine, B., Cuervo, A. M. & Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075 (2008)
Levine, B. & Kroemer, G. Autophagy in the pathogenesis of disease. Cell 132, 27–42 (2008)
Rabinowitz, J. D. & White, E. Autophagy and metabolism. Science 330, 1344–1348 (2010)
Evans, R. M., Barish, G. D. & Wang, Y. X. PPARs and the complex journey to obesity. Nature Med. 10, 355–361 (2004)
Chawla, A., Saez, E. & Evans, R. M. Don’t know much bile-ology. Cell 103, 1–4 (2000)
Nordlie, R. C., Foster, J. D. & Lange, A. J. Regulation of glucose production by the liver. Annu. Rev. Nutr. 19, 379–406 (1999)
Laplante, M. & Sabatini, D. M. mTOR signaling in growth control and disease. Cell 149, 274–293 (2012)
Inoki, K., Kim, J. & Guan, K. L. AMPK and mTOR in cellular energy homeostasis and drug targets. Annu. Rev. Pharmacol. Toxicol. 52, 381–400 (2012)
Hardie, D. G., Ross, F. A. & Hawley, S. A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nature Rev. Mol. Cell Biol. 13, 251–262 (2012)
Settembre, C. & Ballabio, A. Lysosome: regulator of lipid degradation pathways. Trends Cell Biol. http://dx.doi.org/10.1016/j.tcb.2014.06.006 (21 July 2014)
Göttlicher, M., Widmark, E., Li, Q. & Gustafsson, J. A. Fatty acids activate a chimera of the clofibric acid-activated receptor and the glucocorticoid receptor. Proc. Natl Acad. Sci. USA 89, 4653–4657 (1992)
Keller, H. et al. Fatty acids and retinoids control lipid metabolism through activation of peroxisome proliferator-activated receptor-retinoid X receptor heterodimers. Proc. Natl Acad. Sci. USA 90, 2160–2164 (1993)
Ma, K., Saha, P. K., Chan, L. & Moore, D. D. Farnesoid X receptor is essential for normal glucose homeostasis. J. Clin. Invest. 116, 1102–1109 (2006)
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007)
Mizushima, N., Yoshimori, T. & Levine, B. Methods in mammalian autophagy research. Cell 140, 313–326 (2010)
Klionsky, D. J. et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 8, 445–544 (2012)
Maloney, P. R. et al. Identification of a chemical tool for the orphan nuclear receptor FXR. J. Med. Chem. 43, 2971–2974 (2000)
Brown, P. J. et al. Identification of a subtype selective human PPARα agonist through parallel-array synthesis. Bioorg. Med. Chem. Lett. 11, 1225–1227 (2001)
Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 13, 132–141 (2011)
Thoreen, C. C. et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009)
Lee, S. S. et al. Targeted disruption of the α isoform of the peroxisome proliferator- activated receptor gene in mice results in abolishment of the pleiotropic effects of peroxisome proliferators. Mol. Cell. Biol. 15, 3012–3022 (1995)
Sinal, C. J. et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 (2000)
Mizushima, N., Yamamoto, A., Matsui, M., Yoshimori, T. & Ohsumi, Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol. Biol. Cell 15, 1101–1111 (2004)
Komatsu, M. et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 (2005)
Thomas, A. M. et al. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology 51, 1410–1419 (2010)
Claudel, T. et al. Farnesoid X receptor agonists suppress hepatic apolipoprotein CIII expression. Gastroenterology 125, 544–555 (2003)
Chennamsetty, I. et al. Farnesoid X receptor represses hepatic human APOA gene expression. J. Clin. Invest. 121, 3724–3734 (2011)
Seok, S. et al. Transcriptional regulation of autophagy by an FXR–CREB axis. Nature http://dx.doi.org/10.1038/nature13949 (this issue)
Kerr, T. A. et al. Loss of nuclear receptor SHP impairs but does not eliminate negative feedback regulation of bile acid synthesis. Dev. Cell 2, 713–720 (2002)
Wang, L. et al. Redundant pathways for negative feedback regulation of bile acid production. Dev. Cell 2, 721–731 (2002)
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009)
Mizushima, N., Yoshimori, T. & Ohsumi, Y. The role of Atg proteins in autophagosome formation. Annu. Rev. Cell Dev. Biol. 27, 107–132 (2011)
Sun, Z. et al. Deacetylase-independent function of HDAC3 in transcription and metabolism requires nuclear receptor corepressor. Mol. Cell 52, 769–782 (2013)
Feng, D. et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331, 1315–1319 (2011)
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009)
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008)
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002)
Kundu, M. & Thompson, C. B. Autophagy: basic principles and relevance to disease. Annu. Rev. Pathol. 3, 427–455 (2008)
Jin, M., Liu, X. & Klionsky, D. J. SnapShot: Selective autophagy. Cell 152, 368–368 (2013)
Settembre, C. et al. TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 (2011)
Warr, M. R. et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013)
Shoji-Kawata, S. et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature 494, 201–206 (2013)
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012)
McKnight, N. C. et al. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J. 31, 1931–1946 (2012)
Spurr, A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–42 (1969)
Glauert, A. M. Practical Methods in Electron Microscopy 143–144 (North-Holland American Elsevier, 1975)
Reynolds, E. S. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212 (1963)
Acknowledgements
We thank N. Mizushima for the GFP-LC3Tg/+ mice; T. Yoshimori for the mRFP–GFP–LC3 plasmid; M. Komatsu for the Atg7F/F mice; D. Townley and M. Mancini for transmission electron microscopy and confocal microscopy; the members of the Moore laboratory for comments and additional support. Core facilities supported by grants U54 HD-07495-39, P30 DX56338-05A2, P39 CA125123-04 and S10RR027783-01A1. Next-generation sequencing was performed by the Functional Genomics Core of the Penn Diabetes Research Center (DK19525). This work was supported by funding from the Alkek Foundation and the Robert R. P. Doherty Jr-Welch Chair in Science to D.D.M., and R01 DK49780 and DK43806 to M.A.L.
Author information
Authors and Affiliations
Contributions
J.M.L. conceived the project, designed and performed most experiments, interpreted results, and co-wrote the manuscript. M.W. performed animal experiments and participated in discussion of the results. R.X. analysed PPARα and FXR ChIP-seq data, and designed primers for PPARα ChIP-qPCR. K.H.K. performed ChIP assays and molecular cloning. D.F. performed PPARα ChIP-seq. M.A.L. supervised experimental designs. D.D.M. conceived the project, supervised experimental designs, interpreted results, and co-wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
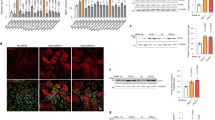
Extended Data Figure 1 PPARα or FXR agonist affects autophagic flux in mouse hepatocytes.
a, Autophagic flux was assessed by LC3 immunoblot analysis in AML12 cells treated with indicated dose of Wy-14,643, or co-treated with both Wy-14,643 and bafilomycin A1 (BafA1). b, c, Biochemical determination of autophagy (p62/SQSTM1 immunoblot) in AML12 cells treated with indicated doses of Wy-14,643 or GW4064 for 24 h. GW4064-treated cells were starved for 2 h. d, Primary hepatocytes from GFP-LC3Tg/+ mice were treated with indicated doses of Torin1 or GW7647. GFP–LC3 cleavage was assessed by GFP immunoblot analysis. e, LC3 immunoblot in AML12 cells treated with indicated doses of GW4064 or co-treated with GW4064 and Torin1. All drug treatments were done in complete media for 24 h unless otherwise indicated. β-actin is a loading control. f, Quantification of autophagic flux shown in Fig. 1a. Numbers of autolysosomes (ALs) induced by 2 h starvation plus vehicle were set as 100%. Numbers of autolysosomes = RFP-positive vesicles – GFP-positive vesicles. Thirty cells were counted per condition (**P < 0.01 versus no starvation plus vehicle; two-tailed t-test). Data represent mean ± s.e.m.
Extended Data Figure 2 Nutrient availability regulates the expression of autophagy-related genes.
a, b, Biochemical determination of autophagy (LC3 immunoblot; LC3-I and LC3-II denote non-lipidated and phosphatidylethanolamine-conjugated forms of MAP1LC3, respectively) or mTORC1 activity (p-S6(Ser 240/244) immunoblot) in AML12 cells treated for indicated time with 100 μM Wy-14,643 or 10 μM GW4064. GW4064-treated cells were starved in HBSS medium for 1 h. β-actin is a loading control. c, Bile acids suppress autophagosome formation. AML12 cells were treated with indicated doses of each bile acid for 24 h, followed by 1-h starvation in HBSS medium. d, Immunoblot analysis of LC3-I/II and β-actin in AML12 cells treated with indicated dose of GW7647 or GW4064 for 24 h. GW4064-treated cells were starved in HBSS medium for 1 h. e, f, Fed ad libitum, 24 h fasted, or 24 h refed after 24 h fasting wild-type mice were euthanized to collect livers at 18:00. e, Immunoblot analysis of LC3-I/II, β-actin, phosphorylated 4E-BP1 (p-4E-BP1(Thr 37/46)) and total 4E-BP1 in liver samples. Proteins (50 μg) from liver homogenates were separated by SDS–PAGE and probed with the indicated antibodies. β-actin is a loading control. A pooled sample was loaded onto the gel in duplicate (n = 5 per group). f, Hepatic expression levels of autophagy-related genes, and PPARα and FXR target genes affected by nutrient availability (n = 5 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice; two-tailed t-test). g, Hepatic expression levels of PPARα or FXR target gene (Acox1 and Cyp7a1, respectively) were determined by qPCR analysis. Fed or fasted wild-type mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day (n = 5 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice). Data represent mean ± s.e.m.
Extended Data Figure 3 Pharmacological activation of PPARα or FXR in fed or fasted mouse liver.
a, c, Hepatic expression levels of autophagy-related genes (LC3a, LC3b and Atg12), PPARα-target gene (Acox1) and FXR-target gene (SHP) were determined by qPCR analysis. Fed or fasted wild-type, Ppara−/− or Fxr−/− mice were orally treated with vehicle, GW7647 or GW4064 twice a day (n = 5 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice treated with vehicle). b, d, Immunoblot analysis of LC3-I/II, β-actin, phosphorylated S6 (p-S6) and total S6 in liver samples. Fed or fasted wild-type mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day. A pooled sample was loaded onto the gel in duplicates (n = 5 per group). β-actin is a loading control. e, Representative confocal images (out of nine tissue sections per condition) of GFP–LC3 puncta (green: autophagosomes) and DAPI (blue: DNA) staining in livers. Fed or fasted bigenic Ppara−/− GFP-LC3Tg/+ or Fxr−/− GFP-LC3Tg/+ mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day. Liver samples were fixed and cryosections were analysed by confocal microscopy. Scale bars, 50 μm. f, Co-localization of BODIPY 493/503 (green) with LC3 (red) in AML12 cells treated with vehicle or 1 μM GW4064 for 24 h and simultaneously cultured with or without 125 μM oleate in complete medium. GW4064-treated cells were starved for 2 h. DNA was stained with DAPI (blue). Scale bars, 20 μm. Quantification of lipophagic vacuoles shown in f and Fig. 3b. Thirty cells were counted per condition (**P < 0.01). g, Measuring β-hydroxybutyrate. AML12 cells were transiently transfected with control siRNA (siControl), Atg5 siRNA (siAtg5) or Atg7 siRNA (siAtg7) for 24 h followed by indicated drug treatments for 48 h with or without 250 μM oleate (vehicle: 0.1% DMSO, Wy: 10 μM Wy-14,643). Released β-hydroxybutyrate in the medium was determined (**P < 0.01 versus siControl treated with vehicle; #P < 0.01 versus siControl treated with Wy-14,643; ##P < 0.01 versus siControl treated with oleate plus Wy-14,643). h, Serum β-hydroxybutyrate were normalized to liver weights. Fed or 24-h fasted control littermates (Atg7F/F) and hepatocyte-specific Atg7F/F-null (Alb-Cre/+ Atg7F/F) mice were treated with vehicle or GW7647 twice a day (n = 4 per group, *P < 0.05, **P < 0.01 versus fed Atg7F/F mice treated with vehicle; #P < 0.01 versus fasted Atg7F/F mice treated with vehicle; ##P < 0.01 versus fasted Atg7F/F mice treated with GW7647). Data are mean ± s.e.m. Statistics by two-tailed t-test.
Extended Data Figure 4 PPARα activation or loss of FXR induces autophagy in liver.
Magnification of representative transmission electron micrograph images (out of 30 cells per group) of livers. a–c, Fed or fasted wild-type, Ppara−/− or Fxr−/− mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day. Lipophagy (yellow arrowheads), autophagosomes (blue arrowheads), autolysosomes (red arrowheads), microautophagy (black arrowheads) and multivesicular bodies (purple arrowheads). Scale bars, 0.5 μm.
Extended Data Figure 5 Expression profiles of autophagy-related genes by PPARα or FXR activation in liver.
a–c, Hepatic expression levels of autophagy-related genes were determined by qPCR analysis in wild-type (a, b), Ppara−/− or Fxr−/− (c) mice. Eleven genes in a were induced by PPARα activation, but not affected by FXR activation. Four genes in b were suppressed by FXR activation, but not affected by PPARα activation (a and b, n = 5 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice treated with vehicle; #P < 0.05, ##P < 0.01 versus fasted wild-type mice treated with vehicle). Altered expression levels of 13 genes shown in Fig. 4a were lost in Ppara−/− or Fxr−/− mice in c (n = 5 per group, *P < 0.05, **P < 0.01 versus fed Ppara−/− or Fxr−/− mice treated with vehicle). Fed or fasted wild-type, Ppara−/− or Fxr−/− mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day. Data represent mean ± s.e.m. Statistics by two-tailed t-test.
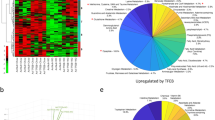
Extended Data Figure 6 Cistromic analysis of PPARα and FXR in mouse liver.
a, De novo motif analysis of PPARα-bound genomic regions. Top PPARα peak regions (±150 base pairs (bp) from peak summits, ranked by enrichment fold) were subjected to de novo motif discovery by MEME. The best motif discovered by MEME (top, E value = 4.7 × 10−227) highly resembles the PPARγ/RXRA heterodimer binding motif from JASPAR (bottom, ID: MA0065.2) as a direct repeat 1 motif (DR1). b, Venn diagram depicting increasing PPARα cistrome upon PPARα agonism in vivo. PPARα highly confident (HC) binding peaks: peaks of wild-type mice treated with vehicle or GW7647 subtracted from peaks of Ppara−/− mice treated with vehicle or GW7647, respectively. c, Venn diagram showing overlapping binding peaks between PPARα ChIP-seq and FXR ChIP-seq from wild-type mice treated with synthetic agonists GW7647 or GW4064. d, Autophagy-related genes of PPARα and FXR cistrome. Within 20 kb from the transcription start site (TSS), PPARα ChIP-seq showed that 7,738 genes of total 28,661 genes (mm9) have highly confident peaks in wild-type mice treated with GW7647 (false discovery rate (FDR) < 0.0001, enrichment over Ppara−/− > 10), and that 124 genes out of 230 autophagy-related genes (HADb: Human Autophagy Database, http://autophagy.lu/) have at least one PPARα peak. FXR ChIP-seq showed that 3,835 genes have peaks in wild-type mice treated with GW4064, and 61 out of 231 autophagy-related genes have at least one FXR peak. e, PPARα ChIP-qPCR for known PPARα target genes (n = 4 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice treated with vehicle; two-tailed t-test). Data represent mean ± s.e.m.
Extended Data Figure 7 PPARα ChIP-seq profiles at loci of autophagy-related genes.
Fed wild-type or Ppara−/− mice were orally gavaged with vehicle or GW7647 twice a day. Mouse livers were taken out 6 h after the last injection of drugs to perform PPARα ChIP-seq and ChIP-qPCR. a, Representative ChIP-seq reads for PPARα aligned to the autophagy-related genes (LC3a, LC3b, Gabarapl1, Bnip3, Atg12, Pex14, Sesn2, Atg7 and Prkaa2). b, PPARα ChIP-qPCR for autophagy-related genes shown in a (n = 4 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice treated with vehicle; two-tailed t-test). Data represent mean ± s.e.m.
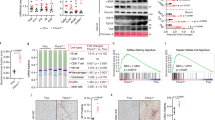
Extended Data Figure 8 PPARα/FXR genomic competition for DR1 in Acox1 gene and autophagy-related genes.
a, Representative ChIP-seq reads for FXR and PPARα aligned to the Acox1 and LC3b genes. The peaks in the box contain DR1 motif. Fed wild-type mice were orally gavaged with vehicle or GW764 twice a day (n = 4 per group). b, PPARα or FXR ChIP-qPCR in livers. Fed or fasted wild-type mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day (n = 3 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice treated with vehicle; ##P < 0.01 versus fasted wild-type mice treated with vehicle). c, Cell-based luciferase reporter assays. AML12 cells were transiently transfected with a 3×PPRE luciferase reporter construct (3×PPRE-luc) and CMX-β-galactosidase in a combination of expression plasmids of PPARα, FXR or both, followed by drug treatment for 20 h (vehicle: 0.1% DMSO; Wy: 10 μM Wy-14,643; GW4064: 1 μM GW4064). Normalized values (luciferase activity/β-galactosidase activity) of vehicle-treated cells transfected with empty plasmid were set as fold 1 (*P < 0.05, **P < 0.01 versus empty treated with vehicle; ##P < 0.01). d, Functional role of DR1 motif in the regulatory region of mouse LC3a and LC3b for PPARα or FXR activity. Cell-based luciferase reporter assays were performed in AML12 cells by transiently transfecting three tandem copies of mouse LC3a/LC3b DR1 luciferase reporter construct (3×LC3a/LC3b DR1 WT-luc) or mutated version (3× LC3a/LC3b DR1 mutant-luc) and CMX-β-galactosidase in a combination of expression plasmids of PPARα, FXR or both, followed by drug treatment for 20 h (vehicle: 0.1% DMSO; Wy: 10 μM Wy-14,643; GW4064: 1 μM GW4064). Normalized values (luciferase activity/β-galactosidase activity) of vehicle-treated cells transfected with empty plasmid were set as fold 1 (**P < 0.01 versus empty treated with vehicle; #P < 0.05, ##P < 0.01). e, Cell-based luciferase reporter assays were performed in AML12 cells by transiently transfecting siControl, siNCoR, siSMRT or siSHP along with three tandem copies of mouse LC3b DR1 luciferase reporter construct (3×LC3b DR1 WT-luc), expression plasmids of PPARα and FXR, and CMX-β-galactosidase followed by drug treatment for 24 h (vehicle: 0.1% DMSO; Wy: 10 μM Wy-14,643; GW4064: 1 μM GW4064). Normalized values (luciferase activity/β-galactosidase activity) of vehicle-treated cells transfected with siControl were set as fold 1 (**P < 0.01 versus siControl treated with vehicle; ##P < 0.01). Data represent mean ± s.e.m. Statistics by two-tailed t-test.
Extended Data Figure 9 PPARα or FXR activation controls recruitments of co-regulators and epigenetic marks in the enhancer regions of LC3a and LC3b genes.
Fed or fasted wild-type mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day. Hepatic ChIP-qPCR analysis with indicated antibodies (p300, NCoR1, SMRT, acetyl-H4 and H3K27me3) was used to determine recruitments of co-regulators and subsequent alterations of epigenetic marks induced by PPARα/FXR genomic competition for DR1 found in the enhancer region of LC3a and LC3b genes (n = 3 per group, *P < 0.05, **P < 0.01 versus fed wild-type mice treated with vehicle; ##P < 0.01 versus fasted wild-type mice treated with vehicle). Data represent mean ± s.e.m. Statistics by two-tailed t-test.
Extended Data Figure 10 Working model of the coordination of hepatic autophagy by nutrient-sensing nuclear receptors, PPARα and FXR.
a, Fed or fasted wild-type mice were orally gavaged with vehicle, GW7647 or GW4064 twice a day. Hepatic CRTC2 ChIP-qPCR in the promoter and enhancer region of LC3a gene (n = 3 per group, **P < 0.01 versus fed wild-type mice treated with vehicle; ##P < 0.01 versus fasted wild-type mice treated with vehicle). Data represent mean ± s.e.m. Statistics by two-tailed t-test. b, Proposed model depicting transcriptionally activating or suppressive nutrient-sensing nuclear receptors, PPARα or FXR, respectively, which coordinate autophagy in liver. Activated PPARα or FXR compete with each other for binding to response elements found in autophagy-related genes.
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-4 and Supplementary Figure 1. (PDF 318 kb)
Supplementary Data
This file contains autophagy-related gene list. (XLSX 43 kb)
Rights and permissions
About this article
Cite this article
Lee, J., Wagner, M., Xiao, R. et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516, 112–115 (2014). https://doi.org/10.1038/nature13961
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13961
This article is cited by
-
PIM1 drives lipid droplet accumulation to promote proliferation and survival in prostate cancer
Oncogene (2024)
-
Emerging role of lipophagy in liver disorders
Molecular and Cellular Biochemistry (2024)
-
Caloric restriction remodels the hepatic chromatin landscape and bile acid metabolism by modulating the gut microbiota
Genome Biology (2023)
-
Transcriptional regulation of autophagy and its implications in human disease
Cell Death & Differentiation (2023)
-
Hepatocyte FBXW7-dependent activity of nutrient-sensing nuclear receptors controls systemic energy homeostasis and NASH progression in male mice
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.