Abstract
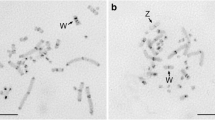
In recent work we have isolated and characterized a highly repetitive DNA (MMV satellite IA) from Muntiacus muntjak vaginalis, the species with the most reduced karyotype in the Cervidae family. We have now analysed the genomes of nine related species for the presence of MMV satellite IA components, and have determined their organization and chromosomal distribution. Repetitive satellite IA type DNA is present in all species of the Cervidae, and also in the bovine, but not in a species of the Tragulidae suggesting that these sequences were generated after the phylogenetic separation of Bovidae and Tragulidae. Studies on the organization of the satellite IA DNA in the various species revealed three main repeat lengths: 1400, 1000 and 807 bp. The relative proportion of satellite IA sequences present in any one of the three registers is strikingly different within the various species and can be correlated with the phylogeny of the Cervidae. The chromosomal locations of the satellite IA sequences were determined in seven species by in situ hybridization. It turned out that the chromosomal rearrangements leading to the reduction in the number of chromosomes during karyotype evolution have led to the elimination of satellite I DNA at most locations. In all tandem fusions, the satellite IA sequences located at the centromeres of the ancestral acrocentric chromosomes are lost. In contrast, during the centric fusion that generates the M. m. vaginalis X chromosome satellite IA sequences are amplified. Sequence motifs, which are known to be involved in recombinational events are present in the satellite IA and might have contributed to the unique karyotype variation in the Cervidae.
Similar content being viewed by others
References
Benedum UM, Neitzel H, Sperling K, Bogenberger JM, Fittler F (1986) Organization and chromosomal distribution of a novel repetitive DNA component from Muntiacus muntjak vaginalis with a repeat length of more than 40 kb. Chromosoma 94:267–272
Bogenberger JM, Schnell H, Fittler F (1982) Characterization of X-chromosome specific satellite DNA of Muntiacus muntjak vaginalis. Chromosoma 87:9–20
Bogenberger JM, Neumaier PS, Fittler F (1985) The Muntjak satellite IA sequence is composed of 31-bp-pair internal repeats that are highly homologous to the 31-base-pair subrepeats of the bovine satellite 1.715. Eur J Biochem 148:55–59
Brinkley BR, Valdivia MM, Tousson A, Brenner SL (1984) Compound kinetochores of the Indian muntac; evolution by linear fusion of kinetochores. Chromosoma 91:1–11
Brutlag DL (1980) Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet 14:121–144
Gaillard C, Doly J, Cortadas J, Bernardi G (1981) The primary structure of bovine satellite 1.715. Nucleic Acids Res 9:6069–6082
Gross-Bellard M, Oudet P, Chambon P (1973) Isolation of highmolecular-weight DNA from mammalian cells. Eur J Biochem 36:32–38
Hörz W, Zachau HG (1977) Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem 73:383–392
Jeffreys AJ, Wilson V, Thein S (1985) Hypervariable minisatellite regions in human DNA. Nature 314:67–73
Johnston FP, Church RB, Lin CC (1982) Chromosome rearrangement between the Indian muntjac and Chinese muntjac is accompanied by a deletion of middle repetitive DNA. Can J Biochem 6:497–506
Liming S, Yingying Y, Xingsheng D (1980) Comparative cytogenetic studies on the red muntjac, Chinese muntjac, and their f1 hybrids. Cytogenet Cell Genet 26:22–27
Maio JJ, Brown FL, Musich PR (1981) Toward a molecular paleontology of primate genomes. Chromosoma 83:103–125
Miklos GLG, John B (1979) Heterochromatin and satellite DNA in man: properties and prospects. Am J Hum Genet 31:264–280
Neitzel H (1982) Karyotypenevolution und deren Bedeutung für den Speciationsprozeß der Cerviden (Cervidae; Artiodactyla; Mammalia). Ph. D. thesis, Fachbereich Biologie der Freien Universität Berlin
Plucienniczak A, Skowronski J, Jaworski J (1982) Nucleotide sequence of bovine 1.715 satellite DNA and its relation to other bovine satellite sequences. J Mol Biol 158:293–304
Rigby PWJ, Dieckmann M, Rhodes C, Berg P (1977) Labelling of deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol 113:237–251
Schmidtke J, Brennecke H, Schmid M, Neitzel H, Sperling K (1981) Evolution of muntjac DNA. Chromosoma 84:187–193
Singer M (1982) Highly repeated sequences in mammalian genomes. Int Rev Cytol 76:67–112
Smith GP (1976) Evolution of repeated DNA seqeunces by unequal crossover. Science 191:528–535
Smith GR, Kunes SM, Schultz DW, Taylor A, Triman KL (1981) Structure of Chi hotspots of generalized recombination. Cell 24:429–436
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Steinmetz M, Stephan D, Lindahl FK (1986) Gene organization and recombinational hotspots in the murine major histocompatibility complex. Cell 44:895–904
Wurster DH, Atkins DB (1972) Muntjac chromosomes: a new karyotype for Muntiacus muntjak. Experientia 28:972–973
Wurster DH, Bernischke K (1970) Indian muntjak Muntiacus muntjak: A deer with a low diploid chromosome number. Science 168:1364–1366
Yu L, Lowensteiner D, Wong EFK, Sawada I, Mazrimas J, Schmid C (1986) Localization and characterization of recombinant DNA clones derived from the highly repetitive DNA sequences in the Indian muntjac cells: Their presence in the Chinese muntjac. Chromosoma 93:521–528
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Bogenberger, J.M., Neitzel, H. & Fittler, F. A highly repetitive DNA component common to all Cervidae: its organization and chromosomal distribution during evolution. Chromosoma 95, 154–161 (1987). https://doi.org/10.1007/BF00332189
Received:
Issue Date:
DOI: https://doi.org/10.1007/BF00332189