Abstract
Objectives
Corynebacterium glutamicum (C. glutamicum) has been harnessed for multi-million-ton scale production of glutamate and lysine. To further increase its amino acid production for fermentation industry, there is an acute need to develop next-generation genome manipulation tool for its metabolic engineering. All reported methods for genome editing triggered with CRISPR-Cas are based on the homologous recombination. While, it requires the generation of DNA repair template, which is a bottle-neck for its extensive application.
Results
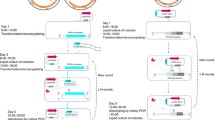
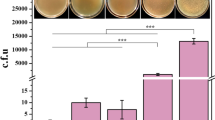
In this study, we developed a method for gene knockout in C. glutamicum via CRISPR-Cpf1-coupled non-homologous end-joining (CC-NHEJ). Specifically, CRISPR-Cpf1 introduced double-strand breaks in the genome of C. glutamicum, which was further repaired by ectopically expressed two NHEJ key proteins (Mycobacterium tuberculosis Ku and ligase D). We provide the proof of concept, for CC-NHEJ, by the successful knockout of the crtYf/e gene in C. glutamicum with the efficiency of 22.00 ± 5.56%, or something like that.
Conclusion
The present study reported a novel genome manipulation method for C. glutamicum.



Similar content being viewed by others
References
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A et al (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157
Bowater R, Doherty AJ (2006) Making ends meet: repairing breaks in bacterial DNA by non-homologous end-joining. Plos Genet 2:93–99
Castaneda-Garcia A, Prieto AI, Rodriguez-Beltran J, Alonso N, Cantillon D, Costas C, Perez-Lago L, Zegeye ED, Herranz M, Plocinski P et al (2017) A non-canonical mismatch repair pathway in prokaryotes. Nat Commun 8:14246
Chen T, Zhu N, Xia H (2014) Aerobic production of succinate from arabinose by metabolically engineered Corynebacterium glutamicum. Bioresour Technol 151:411–414
Chiruvella KK, Liang ZB, Wilson TE (2013) Repair of double-strand breaks by end joining. Csh Perspect Biol 5:a012757
Cho JS, Choi KR, Prabowo CPS, Shin JH, Yang D, Jang J, Lee SY (2017) CRISPR/Cas9-coupled recombineering for metabolic engineering of Corynebacterium glutamicum. Metab Eng 42:157–167
Cromie GA, Connelly JC, Leach DR (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol Cell 8:1163–1174
Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ (2004) Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science (New York, NY) 306:683–685
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551:464–471
Gong CL, Bongiorno P, Martins A, Stephanou NC, Zhu H, Shuman S, Glickman MS (2005) Mechanism of nonhomologous end-joining in mycobacteria: a low-fidelity repair system driven by Ku, ligase D and ligase C. Nat Struct Mol Biol 12:304–312
Heider SA, Peters-Wendisch P, Wendisch VF (2012) Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol 12:198
Ishino S, Skouloubris S, Kudo H, I’Hermitte-Stead C, Es-Sadik A, Lambry JC, Ishino Y, Myllykallio H (2018) Activation of the mismatch-specific endonuclease EndoMS/NucS by the replication clamp is required for high fidelity DNA replication. Nucleic Acids Res 46:6206–6217
Jager W, Schafer A, Puhler A, Labes G, Wohlleben W (1992) Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. J Bacteriol 174:5462–5465
Jiang Y, Qian F, Yang J, Liu Y, Dong F, Xu C, Sun B, Chen B, Xu X, Li Y et al (2017) CRISPR-Cpf1 assisted genome editing of Corynebacterium glutamicum. Nat Commun 8:15179
Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019
Kim HJ, Oh SY, Lee SJ (2020) Single-Base Genome Editing in Corynebacterium glutamicum with the Help of Negative Selection by Target-Mismatched CRISPR/Cpf1. J Microbiol Biotechnol 30:1583–1591
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Krubasik P, Takaichi S, Maoka T, Kobayashi M, Masamoto K, Sandmann G (2001) Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch Microbiol 176:217–223
Krumbach K, Sonntag CK, Eggeling L, Marienhagen J (2019) CRISPR/Cas12a mediated genome editing to introduce amino acid substitutions into the mechanosensitive channel MscCG of Corynebacterium glutamicum. ACS Synth Biol 8:2726–2734
Lee HJ, Lee SJ (2021) Advances in accurate microbial genome-editing CRISPR technologies. J Microbiol Biotechnol 31:903–911
Li L, Wei K, Zheng G, Liu X, Chen S, Jiang W, Lu Y (2018) CRISPR-Cpf1-assisted multiplex genome editing and transcriptional repression in Streptomyces. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00827-18
Lieber MR, Yu K, Raghavan SC (2006) Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair (Amst) 5:1234–1245
Messerotti LJ, Radford AJ, Hodgson AL (1990) Nucleotide sequence of the replication region from the Mycobacterium-Escherichia coli shuttle vector pEP2. Gene 96:147–148
Netzer R, Stafsnes MH, Andreassen T, Goksoyr A, Bruheim P, Brautaset T (2010) Biosynthetic pathway for gamma-cyclic sarcinaxanthin in Micrococcus luteus: heterologous expression and evidence for diverse and multiple catalytic functions of C(50) carotenoid cyclases. J Bacteriol 192:5688–5699
Peng F, Wang X, Sun Y, Dong G, Yang Y, Liu X, Bai Z (2017) Efficient gene editing in Corynebacterium glutamicum using the CRISPR/Cas9 system. Microb Cell Fact 16:201
Sanchez S, Rodríguez-Sanoja R, Ramos A, Demain AL (2017) Our microbes not only produce antibiotics, they also overproduce amino acids. J Antibiot (Tokyo). https://doi.org/10.1038/ja.2017.1142
Sfeir A, Symington LS (2015) Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem Sci 40:701–714
Shuman S, Glickman MS (2007) Bacterial DNA repair by non-homologous end joining. Nat Rev Microbiol 5:852–861
Su T, Liu F, Gu P, Jin H, Chang Y, Wang Q, Liang Q, Qi Q (2016) A CRISPR-Cas9 assisted non-homologous end-joining strategy for one-step engineering of bacterial genome. Sci Rep 6:37895
Sun BB, Yang JJ, Yang S, Ye RD, Chen DJ, Jiang Y (2018a) A CRISPR-Cpf1-assisted non-homologous end joining genome editing system of Mycobacterium smegmatis. Biotechnol J 13:1700588
Sun H, Li F, Liu J, Yang F, Zeng Z, Lv X, Tu M, Liu Y, Ge X, Liu C et al (2018b) A single multiplex crRNA array for FnCpf1-mediated human genome editing. Mol Ther 26:2070–2076
Suzuki N, Nonaka H, Tsuge Y, Inui M, Yukawa H (2005) New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl Environ Microb 71:8472–8480
Tong Y, Charusanti P, Zhang L, Weber T, Lee SY (2015) CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol 4:1020–1029
Wang B, Hu Q, Zhang Y, Shi R, Chai X, Liu Z, Shang X, Zhang Y, Wen T (2018) A RecET-assisted CRISPR-Cas9 genome editing in Corynebacterium glutamicum. Microb Cell Fact. https://doi.org/10.1186/s12934-018-0910-2
Weller GR, Kysela B, Roy R, Tonkin LM, Scanlan E, Della M, Devine SK, Day JP, Wilkinson A, d’Adda di Fagagna F et al (2002) Identification of a DNA nonhomologous end-joining complex in bacteria. Science (New York, NY) 297:1686–1689
Yan MY, Yan HQ, Ren GX, Zhao JP, Guo XP, Sun YC (2017) CRISPR-Cas12a-assisted recombineering in bacteria. Appl Environ Microb. https://doi.org/10.1128/AEM.00947-17
Zahoor A, Lindner SN, Wendisch VF (2012) Metabolic engineering of Corynebacterium glutamicum aimed at alternative carbon sources and new products. Comput Struct Biotechnol J 3:e201210004–e201210004
Zhang J, Yang F, Yang Y, Jiang Y, Huo YX (2019) Optimizing a CRISPR-Cpf1-based genome engineering system for Corynebacterium glutamicum. Microb Cell Fact 18:60
Zhang J, Qian F, Dong F, Wang Q, Yang J, Jiang Y, Yang S (2020) De Novo Engineering of Corynebacterium glutamicum for l-proline production. ACS Synth Biol 9:1897–1906
Zheng X, Li SY, Zhao GP, Wang J (2017) An efficient system for deletion of large DNA fragments in Escherichia coli via introduction of both Cas9 and the non-homologous end joining system from Mycobacterium smegmatis. Biochem Bioph Res Co 485:768–774
Zhu H, Shuman S (2005) A primer-dependent polymerase function of pseudomonas aeruginosa ATP-dependent DNA ligase (LigD). J Biol Chem 280:418–427
Funding
The authors would like to acknowledge financial support of the National Natural Science Foundation of China (No. 31871247), Central Public-interest Scientific Institution Basal Research Fund (No. Y2019PT10, Y2020XK18) and Chinese Academy of Agricultural Sciences grants (No. CAAS-Y2019YJ07-03).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, FY., Wei, N., Zhang, ZH. et al. Genome editing of Corynebacterium glutamicum mediated with Cpf1 plus Ku/LigD. Biotechnol Lett 43, 2273–2281 (2021). https://doi.org/10.1007/s10529-021-03195-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-021-03195-x