Abstract
The delineation of species and their evolutionary relationships informs our understanding of biogeography and how regional faunas are assembled. The peripheral geography and local environment of reefs in the subtropical South Pacific likely promotes the allopatric and adaptive divergence of taxa colonising from the tropics; however, the fauna of this region has been relatively understudied. Here, we address the taxonomic and evolutionary relationships among Chrysiptera taxa of the subtropical South Pacific. We use meristic counts, morphometrics and genetic markers to characterise the similarities and differences among four taxa restricted to the South Pacific region that have strikingly different colouration: C. notialis, a taxon restricted to eastern Australia, New Caledonia, Lord Howe Island and Norfolk Island; C. galba, found in the Cook Islands, southern French Polynesia and Pitcairn Islands; and the two disjunct populations of C. rapanui, found in the eastern Pacific around Rapa Nui (Isla de Pascua or Easter Island) and Motu Motiro Hiva (Salas y Gómez) and in the South-western Pacific around Rangitāhua (Kermadec Islands). Our morphometric analysis confirmed that these four taxa, including the two disjunct populations of C. rapanui, are morphologically distinct. However, our genetic analysis revealed that only C. rapanui from Rapa Nui was genetically differentiated, whereas C. rapanui of Rangitāhua, C. galba and C. notialis all shared a common haplotype. Furthermore, none of the taxa could be consistently differentiated based on individual meristic features. Our study reconciles a formerly perplexing and disjunct distribution for C. rapanui, to reveal that C. rapanui is an endemic of Rapa Nui and that the Chrysiptera of French Polynesia, Rangitāhua, and the South-western Pacific have only a very recent history of divergence. Our analyses suggest these subtropical taxa have diverged from a predominantly tropical Chrysiptera genus in morphological features important in determining colonisation success, locomotion and feeding ecology.
Similar content being viewed by others
Introduction
The Coral Triangle is considered the centre of origin, overlap, accumulation and survival for the shallow marine biodiversity of the Pacific Ocean (reviewed in Bowen et al. 2013; Evans et al. 2016). From the Coral Triangle, there is a southward (Connolly et al. 2003; Mora et al. 2003; Bellwood and Meyer 2009; Tittensor et al. 2010) and eastward (Mora et al. 2003; Bellwood and Meyer 2009; Tittensor et al. 2010; Cowman et al. 2017) decline in species richness across the Pacific, attributed to increasing isolation from the Coral Triangle, a decline in reef area (Bellwood and Hughes 2001; Connolly et al. 2003) and a poleward decrease in resource availability (Gaston 2000; Barneche et al. 2018). None the less, several peripheral regions of the Pacific have a high proportion of endemic species (e.g. Hawai’i, Randall 2007; Rapa Nui, Randall and Cea 2011; Marquesas Islands, Delrieu-Trottin et al. 2015), suggesting their geography, isolation and environment play an important role in the diversification and evolutionary novelty of Pacific-wide biodiversity (Hodge et al. 2012, 2014; Bowen et al. 2013; Cowman et al. 2017).
The subtropical South Pacific consists of several widely spaced island groups that host a relatively depauperate reef fish fauna (Francis 1993; Fig. 1). Generally, fish species richness declines from the west—Australia, Lord Howe Island—into the east—Rapa Nui (Isla de Pascua or Easter Island)—but the proportion of endemism increases across this gradient (Francis 1993; Randall and Cea 2011; Cowman et al. 2017). Few subtropical fishes have broad ranges (Francis 1993); accordingly, the fish fauna of islands in the subtropical South Pacific consists of cosmopolitan tropical species and endemic or range-restricted subtropical species (Francis 1993; Delrieu-Trottin et al. 2018). Although early biogeographic descriptions of the fish fauna suggested that subtropical fish taxa have more restricted ranges in the South-west Pacific (Francis 1993), whereas the fish fauna of South-eastern Pacific was biogeographically cohesive, recent studies have revealed cryptic lineages and endemic species within the South-eastern Pacific region (e.g. Hoese and Stewart 2012; Conway et al. 2018; Delrieu-Trottin et al. 2018). Notably, the closest relatives of several species (and clades) endemic to Rapa Nui are found in Rangitāhua (the Kermadec Islands; Delrieu-Trottin et al. 2018), and not the intervening predominantly tropical islands of French Polynesia. Further examination of the fish fauna across the subtropical South Pacific in its entirety, is required to confidently define the factors that have shaped the diversification of taxa and the resultant biogeography of this relatively understudied region (Briggs and Bowen 2012, 2013).
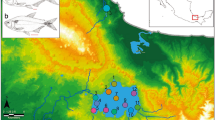
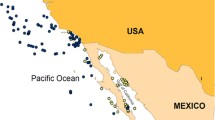
Images and geographic distribution of Chrysiptera taxa included in the study. C. notialis (provided by I. Middleton, Seacologynz.com), C. rapanui (Rangitāhua), C. galba (both provided by I. Skipworth) and C. rapanui (Rapa Nui, provided by L. Rocha, California Academy of Sciences). LI Lord Howe Island, Middleton Reef and Elizabeth Reef; NI Norfolk Island; NC New Caledonia; RK Rangitāhua (Kermadec Islands); French Polynesia including AI Austral Islands, RI Rapa Iti and GI Gambier Islands; PI Pitcairn Island; RN Rapa Nui (Isla de Pascua or Easter Island); MH Motu Motiro Hiva (Salas y Gómez). Circles denote locations of samples included in COI and CytB haplotype networks (see Fig. 5)
Our understanding of the biogeography and the evolutionary history of many Indo-Pacific regional faunas has been enhanced from studying the Pomacentridae. The damselfishes (Pomacentridae) are a major component of the global tropical and temperate ichthyofauna (Allen 1991; Frédérich 2016) and are among the most speciose fish families in these biomes (Cooper et al. 2009). Damselfishes are renowned for their diversity in colour and morphology. Although intraspecific colour variation in relation to geography and local environment is common (Allen 1987, 1991), genetic studies increasingly suggest that these geographic colour variants should be considered separate species (reviewed in Allen et al. 2015b; Victor 2015). In particular, several recent studies focussing on the Chrysiptera genus have described new species within the Coral Triangle and Central Indo-Pacific on the basis of genetic differentiation, meristics and colour (e.g. Allen and Erdmann 2008; Drew et al. 2008, 2010; Allen and Drew 2012; Allen et al. 2015a, b, 2017,2018). These investigations have provided greater understanding of the biogeography and endemism within these regions, and have highlighted the importance of colouration in determining species boundaries within the genus Chrysiptera.
Across the subtropical South Pacific, there are nominally three species of Chrysiptera. Chrysiptera rapanui (Greenfield and Hensley 1970) was described from Rapa Nui, but its distribution was later extended to include Rangitāhua (Allen 1987), on the basis of meristic counts and morphological similarity (Randall and Cea 2011). In the adjacent islands of the South Pacific, C. galba (Allen and Randall 1974) has a relatively broad and continuous distribution across the central South Pacific from the Cook Islands to southern French Polynesia and Pitcairn Islands; and C. notialis (Allen 1975) is distributed in the South-western Pacific from South-eastern Australia to New Caledonia and Norfolk Island. Chrysiptera notialis, C. galba and the geographically disjunct populations of C. rapanui (at Rapa Nui and Rangitāhua) differ considerably in colouration (Fig. 1). The genetic distinction between C. galba and C. rapanui of Rapa Nui was recently confirmed by Delrieu-Trottin et al. (2019) and Tang et al. (2021). However, the genetic affinity of C. rapanui of Rangitāhua and C. notialis relative to these taxa is yet to be assessed. Based on past intuition that geographic colour variants are common within species (Allen 1987, 1991), these subtropical Chrysiptera taxa—C. notialis, C. rapanui of Rangitāhua and C. galba—could plausibly be one species, sister to C. rapanui of Rapa Nui. This hypothesis is supported by their geographic proximity and the described distribution of several other fishes (Briggs and Bowen 2012). Alternatively, as demonstrated by recent descriptions of Chrysiptera species in the Central Indo-Pacific (e.g. Allen et al. 2015a,b, 2017, 2018), all three taxa could be separate species if their colouration is coupled with sufficient morphological and genetic distinction.
Here, we use meristic counts, morphometrics and genetic markers to infer the taxonomic and evolutionary relationships among the subtropical South Pacific Chrysiptera taxa: C. notialis, C. galba and C. rapanui from the disjunct localities of Rapa Nui and Rangitāhua. In using mtDNA genetic markers and meristic measures, we align our examination with a traditional taxonomic investigation, but we additionally consider several morphological measures that are informative regarding ecological and functional divergence among the taxa.
Materials and methods
Taxon sampling
For this study, we made an a priori assumption that the populations of C. rapanui (from Rangitāhua and Rapa Nui) potentially represented two species, and so treated these data separately and hereafter refer to four Chrysiptera taxa: C. notialis, C. galba, C. rapanui from Rapa Nui and C. rapanui from Rangitāhua. To ensure these four taxa were each other's closest relatives, our phylogenetic analysis included all available genetic data for Chrysiptera species (including taxa from neighbouring regions, such as C. starkii; described in “Phylogenetic analyses”).
Expeditions enabled the collection of specimens and tissue samples from New Caledonia (TT in 2017), Norfolk Island (M. Scott), Rangitāhua (DA, LL, TT in 2015), Gambier and Austral Islands (EDT, SP in 2010 and 2014), Pitcairn Island (SP in 2009) and Rapa Nui (EDT, PSA in 2016). A variety of collecting techniques were used (Hawaiian slings, rotenone, clove oil and hand nets; under permits and animal ethics approval to EDT, SP and TT). Whole specimens were preserved in 10% formalin in seawater, and tissues for genetic analysis were preserved in 99% EtOH at ambient temperature.
Additional tissue of C. notialis for genetic analysis was loaned from the Australian Museum. For morphological and meristic measurement, specimens were loaned from several institutions (Auckland Museum, AIM; Australian Museum, Sydney, AMS; Bernice P. Bishop Museum, Hawai’i, BPBM; and California Academy of Sciences, CAS) as follows:
Chrysiptera galba French Polynesia, Tupua’i Lagoon, AMS I.21644–001 (4: 44.3–65.9 mm Standard Length, SL); French Polynesia, Rapa Iti, AMS I.46465–013 (5: 32.1–43.3 mm SL); French Polynesia, Mangareva, AMS I.17348–001 PARATYPE (1: 50.5 mm SL).
C. notialis Australia, Lord Howe Island, AMS I.17376–009 (4: 44.3–61.4 mm SL); Australia, Coral Sea, Elizabeth Reef, AMS I.27149–018 (4: 42.1–81.8 mm SL).
C. rapanui New Zealand, Rangitāhua, Macauley Island, AIM MA655538 (5: 36.0–53.8 mm SL); New Zealand, Rangitāhua, Meyer Island, AIM MA655126 (5: 28.5–43.3 mm SL); Rapa Nui, Motu Tautara, BPBM 39,363 (1: 35.9 mm SL), BPBM 28,164 (1: 30.2 mm SL), BPBM 39,318 (2: 27.0–36.8 mm SL); Rapa Nui, Tahai, BPBM 39,409 (1: 37.8 mm SL); Rapa Nui, Anakana Bay, BPBM 39,206 (4: 27.7–41.6 mm SL); Rapa Nui, Mataveri, BPBM 6689 PARATYPES (4: 20.2–35.1 mm SL); Rapa Nui, Au Akapu, CAS 24690 PARATYPES (4: 30.6–39.8 mm SL).
All information for collected and loaned Chrysiptera samples used in genetic analyses is included in Online Resource 1. Information regarding the Chrysiptera specimens used for morphological measurements and meristic counts is included in Online Resources 2 and 3, respectively.
Morphological and meristic trait measurement and analyses
In total, the morphology of 42 specimens of the genus Chrysiptera were measured for analysis (see Online Resource 2). All measurements were taken from the left side of each fish specimen using digital callipers, to an accuracy of 0.01 mm. Morphometric measurements were (as shown in Fig. 2): anal fin base length (ABL) was the length of the base of the anal fin; body depth (BD) was the vertical distance from the dorsal to ventral body margins aligned with the axis of the pectoral fin base; caudal fin length (CFL) was from the hypural plate margin to the tip of the longest fin ray; caudal peduncle depth (CPD) was the minimum depth of the caudal peduncle; caudal peduncle length (CPL) was the length from the hypural plate margin to the shorter measure of the posterior margin of either the dorsal fin base or the anal fin base; dorsal fin base length (DBL) was the length of the base of the dorsal fin; head length (HL) from the tip of the upper jaw to the posterior margin of the opercle; interorbital width (IOW) was the distance between the dorsal bony margin of the eyes; orbit length (OrL) was the horizontal diameter of the orbit; pectoral fin length (P1L) from the dorsal origin of the fin base to the tip of the longest pectoral fin ray; pelvic fin length (P2L) was from the base of the pelvic spine base to the longest pelvic fin ray; pre-anal length (PAL) was the horizontal distance from the tip of the upper jaw to the base of the first anal spine; pre-dorsal length (PDL) was the horizontal distance from the tip of the upper jaw to the base of the first dorsal spine; pre-orbital length (POL) was from the tip of the upper jaw to the anterior margin of the orbit; pre-pelvic fin length (PPL) was the horizontal distance from the tip of the upper jaw to the base of the pelvic spine; standard length (SL) was from the tip of the upper jaw to the posterior margin of the hypural plate; upper jaw length (UJL) was from the tip of the upper jaw to the posterior margin of the maxilla. Morphological measures for each individual were standardised prior to analysis by dividing each measurement by SL, and thus, our inferences are focused on differences in overall fish shape. To facilitate the interpretation of our results we back-transformed the mean proportions of SL for each taxon to the expected values for a fish of a uniform SL (i.e. SL = 41.2 mm, the mean SL in our study).
Pairwise comparisons of morphological measurements for the Chrysiptera taxa included in the study. Colour indicates how morphological measures differ among taxa, where warm shades indicate that a morphological measure is lesser for the taxon on the x-axis than for the taxon on the y-axis, and cool colours indicate that a morphological measure is greater for the taxon on the x-axis than for the taxon on the y-axis. For instance, the red colour for BD in the comparison of C. galba and C. notialis indicates that C. galba (x-axis) had shallower bodies than C. notialis (y-axis). The intensity of warm and cool colours indicates the strength of the pairwise comparison relative to all comparisons among taxa for that morphological measure. Pairwise significant differences between taxa for a given morphological measure are indicated by a bold box. Inset: illustration of all measured features, except interorbital width (IOW). All morphological measures were analysed as a proportion of standard length (SL); thus, standard length is not included in our analyses of differences in taxon shape
Meristic counts were taken for 39 specimens of the genus Chrysiptera (see Online Resource 3). Meristic counts were obtained with the aid of a stereomicroscope, including pectoral fin rays counted on both left and right sides (P1RL and P1RR, respectively), dorsal fin rays (DoR), dorsal fin spines (DoS) and anal fin rays (AnR). The last ray of the dorsal and anal fin was branched and counted as one ray. Pored lateral line scales (LLP) were counted on both sides of the body posteriorly to the hypural plate margin. The number of scale rows above SALL or below SBLL the lateral line was counted from the origin of the dorsal fin to the lateral line and from lateral line to the origin of the anal fin, respectively. The number of gill rakers were counted (GR) only on the right side.
To examine differences in morphology among the four Chrysiptera taxa we performed one-way ANOVA and Tukey’s HSD tests to quantify differences in mean values of the linear morphological measurements and Fisher’s exact test to determine whether the sampled taxa differed in the proportion of individuals with different counts for the meristic characters using R (version 4.0.2, R Core Team 2020; RStudio version 1.3.1073, RStudio Team 2020). Because many of the morphological measurements and meristic characters were non-independent, we also used linear discriminant analysis (LDA) to explore multivariate differences in morphology. LDA determines the trait combinations that best discriminate among levels of a categorical variable and thus was ideally suited to our dataset where we aimed to find linear combinations of morphological or meristic measurements that discriminated among the four Chrysiptera taxa. For both the morphological measurements and the meristic counts, we used the lda function of the MASS package in R (Venables and Ripley 2002) and leave-one-out cross-validation to determine our prediction accuracy. For the meristic characters, as in canonical correspondence analysis, we χ2 standardised the character indicator matrix before LDA.
Molecular laboratory methods
To conduct our genetic analysis on collected specimens, genomic DNA was extracted using DNeasy Blood and Tissue kits (Qiagen, Valencia, CA), according to the manufacturer’s protocols. To amplify a portion of the COI gene region, we used either the primers FishF2 and FishR2 (described in Ward et al. 2005) or the primer combination named Fish CO1-2 Cocktail (as described in Ivanova et al. 2007), and the Cytochrome B gene region (CytB) was amplified using GluFish-F (Sevilla et al. 2007) and CytbH15573 (Meyer 1993). All PCRs were conducted using MyTaq™ DNA polymerase kits (Bioline, Australia Pty Ltd, Alexandria, NSW), as per the kit instructions. For the Fish CO1-2 primer cocktail, PCR was performed with a denaturation at 94 °C for 1 min, followed by an initial 5 cycles (94 °C for 30 s, 50 °C for 40 secs, 72 °C for 1 min), followed by 35 cycles (94 °C for 30 s, 54 °C for 40 s, 72 °C for 1 min) and then a final extension at 72 °C for 10 min (as described in Ivanova et al. 2007). PCR for CytB was performed with an initial denaturation at 95 °C for 2 min, followed by 35 cycles (95 °C for 45 s, 54 °C for 45 s, 72 °C for 1 min) and then a final extension at 72 °C for 12 min. PCR amplicons were purified using Applied Biosystems™ ExoSAP-IT™ PCR Product Cleanup Reagents and protocol (Thermo Fisher Scientific, West Palm Beach, FL) and sequenced in both directions by Macrogen (Seoul, South Korea) or at the Universidad Austral de Chile’s core facility AUSTRAL-omics (Valdivia, Chile). All sequences are deposited in NCBI (Accessions: KM455353—KM455364, MK657005, MK657007, MK657116, MK657705, MK657836, MK658695, MZ832606—MZ832613, MZ852575—MZ852644) and metadata uploaded to the Genomic Observatories Metadatabase (GEOME, Deck et al. 2017; Riginos et al. 2020; accessioned as Chrysiptera_CYB_CO1_lliggins at https://n2t.net/ark:/21547/Dvj2).
Phylogenetic analyses
The Chrysiptera genus is polyphyletic within the subfamily Pomacentrinae (Quenouille et al 2004; Cooper et al., 2009; Frédérich et al. 2013; McCord et al. 2021; Tang et al. 2021 and references therein). To verify which of the described Chrysiptera clades that each of our focal Chrysiptera taxa have affinity with, and to address the relationship among these taxa, we constructed a phylogeny of Pomacentrinae including our own sequences for C. rapanui (Rapa Nui), C. rapanui (Rangitāhua), C. notialis and C. galba. Although several phylogenies have previously included C. rapanui (Rapa Nui) and C. galba, the origin of the sequences attributed to C. galba has been questioned (see extended discussion of the issue in Tang et al. 2021). Accordingly, Rabosky et al. (2018) discarded C. galba from their phylogeny and Delrieu-Trottin et al. (2019) generated new sequences for C. galba from specimens collected in French Polynesia; these are the sequences we include for C. galba.
We used the regPhylo R package (version 1.0.4; Eme et al. 2019) to query Barcode of Life Database (BOLD, https://www.boldsystems.org/; Ratnasingham et al. 2007) and National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/) for sequences from all other taxa identified within Pomacentrinae according to the World Register of Marine Species (WoRMS, http://www.marinespecies.org/; WoRMS Editorial Board 2021), representatives from the four other Pomacentridae subfamilies (Lepidozyginae, Stegastinae, Abudefdufinae and Chrominae) and other Percomorpha out-group taxa (as per Cooper et al. 2009; Frédérich et al. 2013). From our initial search, the taxa selected for inclusion in the phylogenetic placement of our focal Chrysiptera taxa were species used in previous phylogenies that had less than 50% missing data for the selected gene regions including five mitochondrial (12S, 16S, ND3, COI and CytB) and three nuclear gene regions (RAG1, RAG2 and bmp4).
One representative sequence for each species was chosen based on median sequence length and exported as a fasta file. Newly generated sequence data for our focal Chrysiptera taxa were added to the alignment (COI and CytB). Sequences were aligned using Mafft (Katoh and Standley 2013) using the G-INS-i iterative refinement method which incorporates global pairwise alignment information. Sites with more than 90% gaps were trimmed from alignments using trimAl (Capella-Gutierrez et al. 2009). A maximum likelihood tree was generated using IQ-TREE2 (Minh et al. 2020) from the concatenated gene alignments. The best-fit substitution model was determined for each gene partition and subsequently merged using a greedy strategy until the Bayesian information criterion (BIC) score did not improve using the MFP+ MERGE option in IQ-TREE2 (Kalyaanamoorthy et al. 2017). Node support was inferred with 1000 ultrafast bootstraps generated using the -bb option in IQ-TREE2 (Hoang et al. 2018).
Phylogeographic analyses
To further investigate the genetic and geographic relationships among the focal Chrysiptera taxa, haplotype networks were created for all newly generated and available COI and CytB sequences. For COI, this included: 3 C. rapanui sequences from Rangitāhua, 2 C. rapanui sequences from Rapa Nui, 3 C. galba sequences from Gambier islands, 3 C. galba sequences from Austral islands, 2 C. notialis sequences from Norfolk Island and 1 C. notialis sequence from New Caledonia; and for CytB: 48 C. rapanui sequences from Rangitāhua, 16 C. rapanui sequences from Rapa Nui, 44 C. galba sequences from Gambier islands, 23 C. galba sequences from Pitcairn Island, 3 C. notialis sequences from Norfolk Island and 1 C. notialis sequence from New Caledonia (see Online Resource 1). The haplotype networks were constructed with the program PopArt (Leigh and Bryant 2015) using the TCS algorithm option (Clement et al. 2002) and PopArtHelper (https://github.com/ignacio3437/PopArtHelper) was used to code the locality data into a nexus file that could be loaded into PopArt. The R package ape (Paradis and Schliep 2019) was used to calculate pairwise genetic distances among taxa based on COI and CytB and using the Tamura and Nei (1993) substitution model (selected using the IQ-TREE2 -m TESTONLY function as having the best BIC score).
Results
Morphological analysis
We found strong differences in the measured morphological traits among the four focal Chrysiptera taxa (Fig. 2). Generally, C. notialis had a deeper body but shorter jaw than C. rapanui (Rangitāhua), C. rapanui (Rapa Nui) and C. galba. Chrysiptera rapanui (Rapa Nui) had larger eyes, a longer head, snout and, pre-dorsal as well as pre-pectoral fin lengths, but shorter anal fin base and pelvic fins than C. notialis, C. rapanui (Rangitāhua) and C. galba. Chrysiptera rapanui (Rangitāhua) and C. notialis had narrower interorbital width and longer tails than C. rapanui (Rapa Nui) and C. galba, whereas C. rapanui (Rangitāhua and Rapa Nui) had smaller pectoral fins than C. notialis and C. galba.
Given the number of significant univariate differences in the mean values of the morphological measurements, it is unsurprising that our prediction accuracy for the LDA was excellent—100% prediction accuracy for all taxa given the morphological measurements we examined. This result indicates that each of the four taxa were morphologically distinct with C. notialis being the most different from the other three taxa for the first linear discriminant axis (Fig. 3a). The strong contributions from BD and P1L at one extreme, and UJL and OrL at the other extreme to LDA1 indicated that C. notialis had a deeper body and longer pectoral fins than C. rapanui (Rangitāhua and Rapa Nui) which had larger eyes and longer upper jaw. C. galba was more similar in these features to C. rapanui than to C. notialis. For the second linear discriminant axis, C. rapanui (Rangitāhua) and C. galba were most different with C. rapanui (Rapa Nui) and C. notialis being intermediate for this trait combination. The strong contributions from OrL and POL at one extreme and UJL, PDL and HL at the other extreme to LDA2 indicated that C. rapanui (Rangitāhua) had a different head shape to C. galba. Chrysiptera rapanui (Rangitāhua) had a more elongate head, with longer upper jaw and pre-dorsal length, whereas C. galba had larger eyes and longer snouts (POL). The other two taxa sat intermediate to these extremes, with C. notialis being more similar to C. rapanui (Rangitāhua) and C. rapanui (Rapa Nui) being more similar to C. galba.
Linear discriminant analysis (LDA) of the four Chrysiptera taxa based on morphological measures (a); see Fig. 2 for measured features) and meristic counts (b)
We found no evidence for consistent differences among taxa in the proportions of individuals with different counts for the meristic characters. Examining the multivariate trait combinations (Fig. 3b) that best discriminate among the sampled taxa was more powerful and indicated reasonable prediction accuracy with C. galba, C. notialis and C. rapanui (Rapa Nui) having the lowest prediction accuracy (70%—75%) and C. rapanui (Rangitāhua) having the highest prediction accuracy (90%). The character states contributing most strongly to differences among species in their meristic character counts were the number of scales above (SALL) and below SBLL the lateral line and, to a lesser extent, the number of pectoral fin rays (P1RL, P1RR).
Phylogenetic analysis
The average gene region occupancy for our phylogenetic dataset was 88% for all taxa and 32% for four focal Chrysiptera taxa, with 100% of the most variable sites included (COI and CytB; Online Resource 4). The topology of our phylogeny was similar to previous phylogenies of Pomacentrinae and was well supported (Fig. 4). Accordingly, the Chrysiptera genus was polyphyletic. The four focal Chrysiptera taxa formed a monophyletic clade, sister to C. starcki (corresponding to the Oceanic “Chrysiptera” described by Tang et al. 2021). Chrysiptera rapanui of Rapa Nui was sister to the other three focal Chrysiptera taxa, which were indistinguishable from each other in the phylogeny.
Maximum likelihood phylogenetic tree of the Chrysiptera genus and relatives based on eight gene regions (12S, 16S, ND3, COI, CytB, RAG1, RAG2 and bmp4). The focal Chrysiptera taxa are indicated by the black bar to the right of the phylogeny. Support values were calculated using 1000 ultrafast (UF) bootstraps. Nodes with 100% UF bootstrap support are omitted and some clades are collapsed for clarity
Phylogeographic analyses
The haplotype network based on CytB (729 bp) confirmed that all four Chrysiptera taxa had unique haplotypes, but that C. galba, C. notialis and C. rapanui (Rangitāhua) shared one high-frequency haplotype, from which their unique haplotypes derived (Fig. 5). The haplotypes of C. rapanui (Rapa Nui) were several mutational steps separated from the haplotypes of the other taxa. The COI (585 bp) haplotype network showed a commensurate pattern of divergence, whereby C. rapanui (Rapa Nui) was several mutational steps separated from the other taxa, and C. galba, C. notialis and C. rapanui (Rangitāhua) shared one high-frequency haplotype with few derived haplotypes. On average, sequences of C. rapanui (Rapa Nui) individuals were around 1.65% (based on COI) and 2% (based on CytB) different from the other three taxa (Table 1), whereas all other taxa were less than 0.15% and 0.25% different based on the two gene regions, respectively. The mean level of sequence divergence found within all four focal Chrysiptera taxa was equal to or less than 0.15% for COI and 0.2% for CytB.
Haplotype network based on CytB (a) and COI (b) gene regions of the four Chrysiptera taxa. Each circle represents a unique haplotype. The frequency of each haplotype is indicated by size and the origin of the sequenced specimens is indicated by colour (see key). Edges between haplotypes or small crossbars indicate a mutational step. For sampled locations and their abbreviations see Fig. 1
Discussion
Examining the evolutionary history of closely related damselfishes, including Chrysiptera, has informed our understanding of how geography, and various selective pressures, has driven species evolution across the Indo-Pacific Ocean. The isolated islands and archipelagos of the subtropical South Pacific have been colonised by relatively few damselfish lineages (Cooper et al. 2009), and at these depauperate, poorly connected, high-latitude locations, taxa are likely subjected to different evolutionary pressures than in the tropics. Our analysis of four Chrysiptera taxa with allopatric distributions that span the subtropical South Pacific revealed that they are sister taxa, and have a very recent evolutionary history of divergence, despite their contrasting colouration and morphology. Our study showed that C. rapanui of Rapa Nui in the eastern Central Pacific, should be considered endemic to that region based on genetic, morphological and colour divergence. For the other three taxa examined here, spanning from Pitcairn Island and French Polynesia in the east, to the far western South Pacific, we found limited genetic divergence, suggesting that the divergent morphology and colouration of C. galba, C. notialis and C. rapanui of Rangitāhua (hereafter the Rangitāhua demoiselle) has arisen recently. Our study describes how these subtropical taxa have morphologically diverged from a predominantly tropical Chrysiptera genus. Moreover, our results support other recent work suggesting that the biogeography and speciation patterns in the subtropical South Pacific require further examination (e.g. Hoese and Stewart 2012; Conway et al. 2018; Delrieu-Trottin et al. 2018).
Morphology is often used to inform taxonomy as it is indicative of a species’ ecological function within an environment (Floeter et al. 2018), and the evolutionary pressures the lineage has been subjected to. Our four Chrysiptera taxa were distinguishable based on morphological features previously described as being important in determining the colonisation ability of reef fishes (as per Luiz et al. 2015), their locomotion (Blake 1983) and feeding ecology (Schluter 1993; Cooper and Westneat 2009; Friedman et al. 2020). Overall, C. notialis was the most morphologically distinct taxon, with one of the contributing morphological features being large body depth (relative to standard length). Several studies of reef fishes have described a correlation between body depth (or overall size) of species and colonisation success (Luiz et al. 2015; Jacquet et al. 2017; Ottimofiore et al. 2017). It is thought that the increased body size increases fecundity (Barneche et al. 2018), reduces predation (Claverie and Wainwright 2014; Hodge et al. 2018), increases competitiveness and increases the diversity of food resources consumed (Andersson et al. 2006; Jacquet et al. 2017; Barneche et al. 2018; reviewed in Kulbicki et al. 2015). In our study, C. notialis had a greater body depth (18.25 mm for a fish of 41.2 mm SL, as explained in the Materials and Methods) than the other taxa (with C. galba intermediate [16.23 mm] to C. rapanui [15.48 mm] and the Rangitāhua demoiselle [15.39 mm]), suggesting that this morphological feature has been important in the establishment of their peripheral population and/or maintenance of their meta-population structure, through the (re)colonisation of patchy locations.
Deeper bodies also have implications for the locomotion of fishes (Blake 1983). On the one hand, increased body depth potentially reduces body fineness, increasing form drag (Bainbridge 1960; Hobson 1991). On the other hand, deeper bodies may enhance manoeuvrability in hydrodynamically unstable environments (Webb et al. 1996 and references therein; Friedman et al. 2020) and are often associated with benthic species and ecotypes (e.g. Kusche et al. 2014; Tavera et al. 2018). Potentially to compensate for, or to augment, their deeper body, C. notialis had the longest caudal fin, followed by the Rangitāhua demoiselle (17.01 mm and 16.36 mm, respectively). In the case of C. notialis, a longer caudal fin may enable greater speed and acceleration despite the increased drag created by their greater body depth (Blake 2004), or to supplement their increased manoeuvrability as a consequence of their greater body depth (Webb et al. 1996). For the Rangitāhua demoiselle, a long caudal fin could also provide speed and acceleration, but may also indicate a tendency towards steady locomotion in mid-water (Randall 1967; Hobson 1991; Friedman et al. 2020). When under threat of predation, reef fishes will either seek shelter where it is available or attempt to outswim the predator. In rocky reef environments with low coral cover, adequate reef structure to provide shelter to damselfishes is limited or absent (Anderson et al. 1989). The Rangitāhua demoiselle does not have a strong association with the benthos, instead, forming large schools in the water column above shallow reef areas (L. Liggins). This habit contrasts with that described for C. galba, for example (S. Planes) where individuals are associated with the benthos. These differing habits, and water column positioning, suggest that the ecological function of these taxa may differ.
Fish body shape changes associated with transitions between pelagic and benthic habits are well described (Friedman et al. 2020 and references therein). Damselfishes are particularly renowned for their labile transitions among dietary ecotypes along a bentho-pelagic (or limnetic) niche axis (Cooper et al. 2017; McCord et al. 2021). Species vary from being pelagic feeders that sit higher in the water column and feed on plankton, to benthic feeders that maintain a close association with the benthos feeding on algae and detritus (Wilson and Bellwood 1997), and omnivorous, intermediary, bentho-pelagic feeders. Several studies suggest that fishes with deeper bodies are more efficient benthic feeders, whereas more slender species are superior at pelagic feeding (Davis and Birdsong 1973; Hobson 1991; Friedman et al. 2020; e.g. Schluter 1993; Kusche et al. 2014; Tavera et al. 2018). Based on our study, C. notialis would be presumed to be a better benthic feeder (based on their greater body depth), whereas the more slender body shapes of C. rapanui and the Rangitāhua demoiselle make them more efficient plankton feeders. Accordingly, in a recent review of Pomacentridae phylogenetics and dietary ecotype, McCord et al. (2021) describe C. galba as bentho-pelagic and C. rapanui as pelagic (the Rangitāhua demoiselle and C. notialis were not examined). Their comparative phylogenetic analysis suggested a more frequent transition from bentho-pelagic to pelagic dietary ecotypes in derived lineages of damselfishes (McCord et al. 2021; also suggested by Floeter et al. 2018; Gajdzik et al. 2019).
Several studies have associated changes in damselfish jaw and head (cranial) morphology with a species position on the bentho-pelagic niche axis (Cooper and Westneat 2009; Frédérich et al. 2013; Cooper et al. 2017). In our study, C. rapanui had the longest upper jaw, followed by the Rangitāhua demoiselle (relative to standard length). This trait (along with the ability to protrude its jaw) has been associated with efficacy in pelagic feeding in damselfishes (Cooper et al. 2017), suggesting that C. rapanui and the Rangitāhua demoiselle are more planktivorous than C. notialis and C. galba. This inference based on studies of damselfishes contrasts with findings in several other fishes, where large jaw and mouth features are associated with benthic feeding (e.g. haemulids, Tavera et al. 2018; Friedman et al. 2020), small mouths with planktonic feeding (Davis and Birdsong 1973) and small mouths with equal feeding efficiency in both benthic and pelagic habits (Kotrschal 1988; Hobson 1991). In our own study, the longer upper jaw in the exclusively subtropical taxa (the Rangitāhua demoiselle and C. rapanui) compared with the tropical–subtropical taxa (C. galba and C. notialis) would support a transition to a pelagic feeding mode in derived lineages (as in McCord et al. 2021), but is also potentially influenced by a tendency towards planktivorous feeding at higher latitude, less oligotrophic locations (Holland et al. 2020).
Comparisons among our four focal Chrysiptera taxa also revealed that C. rapanui (Rapa Nui) had the largest eyes (4.57 mm) followed by C. galba (4.43 mm). Increased orbit length may implicate a diet shift towards smaller or more mobile prey where larger eyes allow for more precise capture, potentially important in both planktonic and benthic feeding (Davis and Birdsong 1973; Goatley and Bellwood 2009). As such, this morphological feature cannot be directly associated with the bentho-pelagic axis, and its relative measurement is influenced by other features that would be expected to vary along this axis (see Davis and Birdsong 1973). In summary, when compared with C. rapanui and the Rangitāhua demoiselle, C. galba and C. notialis both had a short head and short jaws, as well as deeper bodies and longer pectoral fins important in increasing manoeuvrability in benthic labriform swimming. Definitive support for these associations of morphology with ecological function for our examined Chrysiptera taxa requires comparative analysis of diet, observation of feeding strategies including jaw mechanics and ecological survey of bentho-pelagic positioning and predator response. Nonetheless, rapid evolution of jaw and head morphology, as well as body depth, in response to diet is commonly described in pomacentrids (e.g. Frédérich et al. 2008, 2013; Cooper and Westneat 2009). Together with anecdotal behavioural observations, these differences in morphology do support a transition to a more pelagic existence of Chrysiptera taxa in the subtropical South Pacific.
Although our morphological analysis suggests that the four Chrysiptera taxa are on different evolutionary trajectories, there was little genetic differentiation among the taxa and this did not correlate with patterns of morphological differentiation. Genetic differentiation based on mitochondrial markers presumably reflects the neutral processes of migration, mutation and drift wherein the amount of genetic differentiation roughly scales with time. Several studies of coral reef fishes have revealed concordance among such genetic divergence and morphological divergence (e.g. Rocha 2004; Drew et al. 2008, 2010). Nonetheless, similar to our findings, a recent study of haemulid fishes found that the level of divergence in phenotypic traits and colour of sister species, including those that occur in allopatry, was uncorrelated with time since divergence (Tavera and Wainwright 2019). Furthermore, studies focused on individuals within fish populations have suggested that morphological change and associated diet specialisation occurs at an early ecological and evolutionary stage of divergence, and before lineage diversification (Kusche et al. 2014).
In our study, the levels of sequence divergence at CytB and COI among the four focal taxa were less than typically observed between fish species (e.g. Ward et al. 2005), including for recently described Chrysiptera species (e.g. Allen et al. 2015a, b, where sequence divergence was > 3%; Table 1). Nonetheless, C. rapanui was genetically differentiated and genetically distinct from the other taxa (Table 1, Figs. 4 and 5), indicating this taxon has been isolated from gene flow for the longest period of time and should have distinct species status as an endemic of Rapa Nui. Rapa Nui is the most isolated subtropical island of the South Pacific; a fact often related to it also having a high proportion of endemic species (almost 22%, Randall and Cea 2011). Our findings support other recent molecular studies suggesting this level of endemism may be higher than previously described (Delrieu-Trottin et al. 2018, 2019).
Despite the small size and isolation of the island groups inhabited by the Rangitāhua demoiselle and C. notialis (distribution is centred on Norfolk Island and Lord Howe Island) in the subtropical South-west Pacific, these taxa were not as genetically differentiated as C. rapanui. Chrysiptera galba, C. notialis and the Rangitāhua demoiselle each had unique haplotypes indicative of restricted contemporary gene flow; however, these haplotypes were only distinguished by a few mutations, and the taxa also shared one haplotype. This ancestral haplotype likely persists through incomplete lineage sorting, whereby random drift has not had enough time to “sort” this haplotype from the large meta-population. Such incomplete lineage sorting (also described for shallow marine invertebrates of Rangitāhua, Liggins et al. 2014) and the generally low endemism of the fish fauna found at Rangitāhua, Norfolk Island and Lord Howe Island (each < 5%, Francis 1993, 2019; Francis and Duffy 2015) are likely a consequence of their recent connectivity with each other and to neighbouring regions. Furthermore, although emergent landmasses have likely been present throughout the South-west Pacific since the Pliocene (summarised in Francis 1993), and recent studies have revealed paleoendemism (sensu Stebbins and Major 1965; e.g. Bronstein et al. 2017), the present configuration of the islands is relatively recent (Rangitāhua, 2.5 Ma, Brook 1989; Norfolk < 3.1 Ma, Jones and McDougall 1973; Lord Howe, < 6.9 Ma, McDougall et al. 1981). Accordingly, the molecular biogeography (sensu Crandall et al. 2019) of the examined South-west–Central Pacific Chrysiptera taxa (C. notialis, C. galba and the Rangitāhua demoiselle) supports the biogeographic clustering of Lord Howe Island, Norfolk Island, Rangitāhua and French Polynesia separate from the far eastern Central Pacific (as in the Provinces proposed by Briggs and Bowen 2012, and Cowman et al. 2017).
It is well established that the processes that generate species operate over both ecological and evolutionary timescales, and as such, the characterisation of species divergence benefits from multiple lines of investigation (Simpson 1944; Reznik and Ricklefs 2009). Here, we examined the genetic and morphological divergence of Chrysiptera taxa of the subtropical South Pacific. We found some evidence for considering each taxon as a distinct species—namely colour, morphology and the sharing of just one CytB haplotype. Yet, traditional meristic features could not reliably distinguish among the taxa. Using a multivariate approach, we had the greatest ability to distinguish the Rangitāhua demoiselle based on meristic measures (90% classification success, Fig. 3b). On this basis, we intend to describe the Rangitāhua demoiselle as a unique species, endemic to Rangitāhua. It is likely that a genomic approach, such as using RAD-seq or Ultra Conserved Elements (described by Smith et al. 2014), would provide greater resolution regarding the evolutionary relationships among these taxa (as in DiBattista et al. 2018; Stiller et al. 2021) and would help to resolve their taxonomy (as in Bernardi et al. 2017; Erickson et al. 2021; Tea et al. 2021).
The relative roles of geographic and biological isolating mechanisms among taxa can vary over the course of speciation (Norris and Hull 2012). In the current changing climate, wherein we anticipate altered dispersal patterns (Lett et al. 2010) and increased colonisation of higher latitude regions from the tropics (Molinos et al. 2016; Liggins et al. 2020), it remains to be seen whether the current allopatry of these Chrysiptera taxa persists. In temperate, mainland Aotearoa New Zealand, stray C. notialis and the Rangitāhua demoiselle co-occur within a few kilometres of each other (Francis et al. 1999; Middleton et al. 2021); in this setting, their ecological function and any evolution of traits associated with reproduction will have the greatest bearing on their future evolutionary trajectories.
References
Allen GR (1987) A new species of pomacentrid fish with notes on other damselfishes of the Kermadec Islands. Rec West Aust Mus 13:263–273
Allen GR (1991) Damselfishes of the World. Mergus Press, Germany, Melle
Allen GR, Randall JE (1974) Five new species and a new genus of damselfishes (Family Pomacentridae) from the South Pacific Ocean. Trop Fish Hobbyist 21:36–46, 58–49
Allen GR, Drew JA (2012) A new species of damselfish (Pomacentridae) from Fiji and Tonga. Aqua 18:171–180
Allen GR, Erdmann MV (2008) A new species of damselfish (Pomacentridae: Chrysiptera) from western New Guinea and the Togean Islands, Indonesia. Aqua 13:171–178
Allen GR, Erdmann MV, Cahyani NKD (2015a) Review of the Chrysiptera oxycephala complex of damselfishes (Pomacentridae) with descriptions of three new species from the East Indian Archipelago. J Ocean Sci Found 17:56–84
Allen GR, Erdmann MV, Kurniasih EM (2015b) Chrysiptera caesifrons, a new species of damselfish (Pomacentridae) from the south–western Pacific Ocean. J Ocean Sci Found 15:16–32
Allen GR, Erdmann MV, Cahyani DNK (2017) A new species of damselfish (Chrysiptera: Pomacentridae) from coral reefs of the Solomon Islands. J Ocean Sci Found 28:10–21
Allen GR, Erdmann MV, Cahyani DNK (2018) Chrysiptera uswanasi, a new microendemic species of damselfish (Teleostei: Pomacentridae) from West Papua Province, Indonesia. J Ocean Sci Found 31:74–86
Anderson TW, DeMartini EE, Roberts DA (1989) The relationship between habitat structure, body size and distribution of fishes at a temperate artificial reef. Bull Mar Sci 44:681–697
Andersson J, Johansson F, Söderlund T (2006) Interactions between predator- and diet-induced phenotypic changes in body shape of crucian carp. Proc Roy Soc Lond B Biol Sci 273:431–437
Bainbridge R (1960) Speed and stamina in three fish. J Exp Biol 37:129–153
Barneche DR, Robertson DR, White CR, Marshall DJ (2018) Fish reproductive-energy output increases disproportionately with body size. Science 360:642–645
Bellwood DR, Hughes TP (2001) Regional-scale assembly rules and biodiversity of coral reefs. Science 292:1532–1535
Bellwood DR, Meyer CP (2009) Searching for heat in a marine biodiversity hotspot. J Biogeogr 36:569–576
Bernardi G, Longo GC, Quiros TAL (2017) Altrichthys alelia, a new brooding damselfish (Teleostei, perciformes, pomacentridae) from Busuanga Island. Philippines Zookeys 675:45
Blake RW (1983) Fish locomotion. Cambridge University Press, United Kingdom, Cambridge
Blake RW (2004) Fish functional design and swimming performance. J Fish Biol 65:1193–1222
Bowen BW, Rocha LA, Toonen RJ, Karl SA, Laboratory ToBo (2013) The origins of tropical marine biodiversity. TREE 28:359–366
Briggs JC, Bowen BW (2012) A realignment of marine biogeographic provinces with particular reference to fish distributions. J Biogeogr 39:12–30
Briggs JC, Bowen BW (2013) Marine shelf habitat: biogeography and evolution. J Biogeogr 40:1023–1035
Bronstein O, Kroh A, Tautscher B, Liggins L, Haring E (2017) Cryptic speciation in pan-tropical sea urchins: a case study of an edge-of-range population of Tripneustes from the Kermadec Islands. Sci Rep 7:1–16
Brook FJ (1989) Recent shallow marine scleractinian corals at subtropical Kermadec Islands, Southwest Pacific. Report held by New Zealand Department of Conservation, Auckland
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) Trim Al: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973
Claverie T, Wainwright PC (2014) A morphospace for reef fishes: elongation is the dominant axis of body shape evolution. PLoS ONE 19:e112732
Clement M, Snell Q, Walker P, Posada D, Crandall K (2002) TCS: estimating gene genealogies. In: Parallel and Distributed Processing Symposium 3: 0184. IEEE Computer Society
Connolly SR, Bellwood DR, Hughes TP (2003) Indo-Pacific biodiversity of coral reefs: deviations from a mid-domain model. Ecology 84:2178–2190
Conway KW, Stewart AL, Summers AP (2018) A new genus and species of clingfish from the Rangitāhua Kermadec Islands of New Zealand (Teleostei, Gobiesocidae). Zookeys 786:75
Cooper WJ, Westneat MW (2009) Form and function of damselfish skulls: rapid and repeated evolution into a limited number of trophic niches. BMC Evol Biol 9:1–17
Cooper WJ, Smith LL, Westneat MW (2009) Exploring the radiation of a diverse reef fish family: phylogenetics of the damselfishes (pomacentridae), with new classifications based on molecular analyses of all genera. Mol Phylo Evol 52:1–16
Cooper WJ, Carter CB, Conith AJ, Rice AN, Westneat MW (2017) The evolution of jaw protrusion mechanics is tightly coupled to bentho-pelagic divergence in damselfishes (Pomacentridae). J Exp Biol 220:652–666
Cowman PF, Parravicini V, Kulbicki M, Floeter SR (2017) The biogeography of tropical reef fishes: endemism and provinciality through time. Biol Rev 92:2112–2130
Crandall ED, Riginos C, Bird CE, Liggins L, Treml E, Beger M, Barber PH, Connolly SR, Cowman PF, DiBattista JD, Eble JA (2019) The molecular biogeography of the Indo-Pacific: testing hypotheses with multispecies genetic patterns. Glob Ecol Biogeogr 28:943–960
Davis WP, Birdsong RS (1973) Coral reef fishes which forage in the water column. Helgoländer Wiss Meeresunters 24:292–306
Deck J, Gaither MR, Ewing R, Bird CE, Davies N, Meyer C, Riginos C, Toonen RJ, Crandall ED (2017) The genomic observatories metadatabase (GeOMe): a new repository for field and sampling event metadata associated with genetic samples. PLoS Biol 15:e2002925
Delrieu-Trottin E, Williams JT, Bacchet P, Kulbicki M, Mourier J, Galzin R, de Loma TL, Mou-Tham G, Siu G, Planes S (2015) Shore fishes of the Marquesas Islands, an updated checklist with new records and new percentage of endemic species. Check List 11:1758
Delrieu-Trottin E, Liggins L, Trnski T, Williams JT, Neglia V, Rapu-Edmunds C, Planes S, Saenz-Agudelo P (2018) Evidence of cryptic species in the blenniid Cirripectes alboapicalis species complex, with zoogeographic implications for the South Pacific. Zookeys 810:127
Delrieu-Trottin E, Brosseau-Acquaviva L, Mona S, Neglia V, Giles EC, Rapu-Edmunds C, Saenz-Agudelo P (2019) Understanding the origin of the most isolated endemic reef fish fauna of the Indo-Pacific: coral reef fishes of Rapa Nui. J Biogeogr 46:723–733
DiBattista JD, Alfaro ME, Sorenson L, Choat JH, Hobbs JPA, Sinclair-Taylor TH, Rocha LA, Chang J, Luiz OJ, Cowman PF, Friedman M (2018) Ice ages and butterflyfishes: phylogenomics elucidates the ecological and evolutionary history of reef fishes in an endemism hotspot. Ecol Evol 22:10989–11008
Drew JA, Allen GR, Kaufman L, Barber PH (2008) Endemism and regional color and genetic differences in five putatively cosmopolitan reef fishes. Conserv Biol 22:965–975
Drew JA, Allen GR, Erdmann MV (2010) Congruence between mitochondrial genes and color morphs in a coral reef fish: population variability in the Indo-Pacific damselfish Chrysiptera rex (Snyder, 1909). Coral Reefs 29:439–444
Eme D, Anderson MJ, Struthers CD, Roberts CD, Liggins L (2019) An integrated pathway for building regional phylogenies for ecological studies. Glob Ecol Biogeogr 28:1899–1911
Erickson KL, Pentico A, Quattrini AM, McFadden CS (2021) New approaches to species delimitation and population structure of anthozoans: two case studies of octocorals using ultraconserved elements and exons. Mol Ecol Res 21:78–92
Evans SM, McKenna C, Simpson SD, Tournois J, Genner MJ (2016) Patterns of species range evolution in Indo-Pacific reef assemblages reveal the Coral Triangle as a net source of transoceanic diversity. Biol Lett 30:20160090
Floeter SR, Bender MG, Siqueira AC, Cowman PF (2018) Phylogenetic perspectives on reef fish functional traits. Biol 93:131–151
Francis MP (1993) Checklist of the coastal fishes of Lord Howe, Norfolk, and Kermadec Islands, Southwest Pacific Ocean. Pac Sci 47:136–170
Francis MP (2019) Checklist of the coastal fishes of Lord Howe, Norfolk and Kermadec Islands, southwest Pacific Ocean. Figshare: https://doi.org/10.6084/m9.figshare.c.4428305.v2
Francis MP, Duffy CA (2015) New records, checklist and biogeography of Kermadec Islands’ coastal fishes. Bull Auck Mus 20:481–495
Francis MP, Worthington CJ, Saul P, Clements KD (1999) New and rare tropical and subtropical fishes from northern New Zealand. NZ J Mar Fresh Res 33:571–586
Frédérich B, Parmentier E (2016) Biology of damselfishes. CRC Press, USA
Frédérich B, Pilet A, Parmentier E, Vandewalle P (2008) Comparative trophic morphology in eight species of damselfishes (Pomacentridae). J Morphol 269:175–188
Frédérich B, Sorenson L, Santini F, Slater GJ, Alfaro ME (2013) Iterative ecological radiation and convergence during the evolutionary history of damselfishes (Pomacentridae). Am Nat 181:94–113
Friedman ST, Price SA, Corn KA, Larouche O, Martinez CM, Wainwright PC (2020) Body shape diversification along the benthic-pelagic axis in marine fishes. Proc R Soc B Biol Sci 287:20201053
Gajdzik L, Aguilar-Medrano R, Frédérich B (2019) Diversification and functional evolution of reef fish feeding guilds. Ecol Lett 22:572–582
Gaston KJ (2000) Global patterns in biodiversity. Nature 405:220–227
Goatley CH, Bellwood DR (2009) Morphological structure in a reef fish assemblage. Coral Reefs 28:449–457
Greenfield DW, Hensley DA (1970) Damselfishes (Pomacentridae) of Easter Island, with descriptions of two new species. Copeia 4:689
Hoang DT, Chernomor O, Von Haeseler A, Minh BQ, Vinh LS (2018) UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522
Hobson ES (1991) Trophic relationships of fishes specialized to feed on zooplankters above coral reefs. In: Sale PF (ed) The Ecology of Fishes on Coral Reefs San Diego. California, Academic Press, USA, pp 69–95
Hodge JR, Read CI, Van Herwerden L, Bellwood DR (2012) The role of peripheral endemism in species diversification: evidence from the coral reef fish genus Anampses (Family: Labridae). Mol Phylo Evol 62:653–663
Hodge JR, Van Herwerden L, Bellwood DR (2014) Temporal evolution of coral reef fishes: global patterns and disparity in isolated locations. J Biogeogr 41:2115–2127
Hodge JR, Alim C, Bertrand NG, Lee W, Price SA, Tran B, Wainwright PC (2018) Ecology shapes the evolutionary trade-off between predator avoidance and defence in coral reef butterflyfishes. Ecol Lett 21:1033–1042
Hoese DF, Stewart AL (2012) A new species of the gobiid genus Eviota (Teleostei: Gobioidei) from the Kermadec Islands, New Zealand. Zootaxa 3418:61–67
Holland MM, Smith JA, Everett JD, Vergés A, Suthers IM (2020) Latitudinal patterns in trophic structure of temperate reef-associated fishes and predicted consequences of climate change. Fish Fish 21:1092–1108
Ivanova NV, Zemlak TS, Hanner RH, Hebert PD (2007) Universal primer cocktails for fish DNA barcoding. Mol Ecol Notes 7:544–548
Jacquet C, Mouillot D, Kulbicki M, Gravel D (2017) Extensions of island biogeography theory predict the scaling of functional trait composition with habitat area and isolation. Ecol Lett 20:135–146
Jones JG, McDougall I (1973) Geological history of Norfolk and Philip Islands, southwest Pacific Ocean. J Geol Soc Aust 20:239–254
Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS (2017) Model Finder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software v. 7. improvements in performance and usability. Mol Biol Evol 30:772–780
Kotrschal K (1988) Evolutionary patterns in tropical marine reef fish feeding. J Zool Syst Evol Res 26:51–64
Kulbicki M, Parravicini V, Mouillot D (2015) Patterns and processes in reef fish body size. In: Mora C (ed) Ecology of fishes on coral reefs. Cambridge University Press, Cambridge
Kusche H, Recknagel H, Elmer KR, Meyer A (2014) Crater lake cichlids individually specialize along the benthic-limnetic axis. Ecol Evol 4:1127–1139
Leigh JW, Bryant D (2015) popart: full-feature software for haplotype network construction. Methods Ecol Evol 6:1110–1116
Lett C, Ayata SD, Huret M, Irisson JO (2010) Biophysical modelling to investigate the effects of climate change on marine population dispersal and connectivity. Prog Oceanogr 87:106–113
Liggins L, Gleeson L, Riginos C (2014) Evaluating edge–of–range genetic patterns for tropical echinoderms, Acanthaster planci and Tripneustes gratilla, of the Kermadec Islands, southwest Pacific. Bull Mar Sci 90:379–397
Liggins L, Sweatman JA, Trnski T, Duffy CA, Eddy TD, Aguirre JD (2020) Natural history footage provides new reef fish biodiversity information for a pristine but rarely visited archipelago. Sci Rep 21:1–7
Luiz OJ, Mendes TC, Barneche DR, Ferreira CG, Noguchi R, Villaça RC, Rangel CA, Gasparini JL, Ferreira CE (2015) Community structure of reef fishes on a remote oceanic island (St Peter and St Paul’s Archipelago, equatorial Atlantic): the relative influence of abiotic and biotic variables. Mar Fresh Res 66:739–749
McCord CL, Cooper WJ, Nash CM, Westneat MW (2021) Phylogeny of the damselfishes (Pomacentridae) and patterns of asymmetrical diversification in body size and feeding ecology. bioRxiv 2021.02.07.430149: https://doi.org/10.1101/2021.02.07.430149
McDougall I, Embleton BJ, Stone DB (1981) Origin and evolution of Lord Howe Island, southwest Pacific Ocean. J Geol Soc Aust 28:155–176
Meyer A (1993) Phylogenetic relationships and evolutionary processes in East African cichlid fishes. TREE 8:279–284
Middleton I, Aguirre JD, Trnski T, Francis M, Duffy C, Liggins L (2021) Introduced alien, range extension or just visiting? Combining citizen science observations and expert knowledge to classify range dynamics of marine fishes. Divers Distrib 27:1278–1293
Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R (2020) IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic Era. Mol Biol Evol 37:1530–1534
Molinos JG, Halpern BS, Schoeman DS, Brown CJ, Kiessling W, Moore PJ, Pandolfi JM, Poloczanska ES, Richardson AJ, Burrows MT (2016) Climate velocity and the future global redistribution of marine biodiversity. Nat Clim Change 6:83–88
Mora C, Chittaro PM, Sale PF, Kritzer JP, Ludsin SA (2003) Patterns and processes in reef fish diversity. Nature 421:933–936
Norris RD, Hull PM (2012) The temporal dimension of marine speciation. Evol Ecol 26:393–415
Ottimofiore E, Albouy C, Leprieur F, Descombes P, Kulbicki M, Mouillot D, Parravicini V, Pellissier L (2017) Responses of coral reef fishes to past climate changes are related to life-history traits. Ecol Evol 7:1996–2005
Paradis E, Schliep K (2019) Ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 35:526–528
Quenouille B, Bermingham E, Planes S (2004) Molecular systematics of the damselfishes (Teleostei: Pomacentridae): bayesian phylogenetic analyses of mitochondrial and nuclear DNA sequences. Mol Phylogenet Evol 31:66–88
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Rabosky DL, Chang J, Title PO, Cowman PF, Sallan L, Friedman M, Kaschner K, Garilao C, Near TJ, Coll M, Alfaro ME (2018) An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559:392–395
Randall JE (1967) Food habits of reef fishes of the West Indies. Stud Trop Oceanogr 5:665–847
Randall JE (2007) Reef fishes of Hawai’i. University of Hawai’i Press, Honolulu
Randall JE, Cea A (2011) Shore fishes of Easter Island. University of Hawai’i Press, Honolulu
Ratnasingham S, Hebert PDN (2007) BOLD: the barcode of life data system. Mol Ecol Notes 7:355–364
Reznick DN, Ricklefs RE (2009) Darwin’s bridge between microevolution and macroevolution. Nature 457:837–842
Riginos C, Crandall ED, Liggins L, Gaither MR, Ewing RB, Meyer C, Andrews KR, Euclide PT, Titus BM, Therkildsen NO, Salces-Castellano A (2020) Building a global genomics observatory: using GEOME (the Genomic Observatories Metadatabase) to expedite and improve deposition and retrieval of genetic data and metadata for biodiversity research. Mol Ecol Res 20:1458–1469
Rocha LA (2004) Mitochondrial DNA and color pattern variation in three western Atlantic Halichoeres (Labridae), with the revalidation of two species. Copeia 4:770–782
RStudio Team (2020) RStudio: Integrated Development for R. RStudio, PBC, Boston. http://www.rstudio.com/
Schluter D (1993) Adaptive radiation in sticklebacks: size, shape, and habitat use efficiency. Ecology 74:699–709
Sevilla RG, Diez A, Norén M, Mouchel O, Jérôme M, Verrez-Bagnis V, Van Pelt H, Favre-Krey LA, Krey G, Consortium TF, Bautista JM 2007 Primers and polymerase chain reaction conditions for DNA barcoding teleost fish based on the mitochondrial cytochrome b and nuclear rhodopsin genes. Mol Ecol Notes,7:730–734
Simpson GG (1944) Tempo and mode in evolution. Columbia University Press, New York
Smith BT, Harvey MG, Faircloth BC, Glenn TC, Brumfield RT (2014) Target capture and massively parallel sequencing of ultraconserved elements for comparative studies at shallow evolutionary time scales. Syst Biol 63:83–95
Stebbins GL, Major J (1965) Endemism and speciation in the California flora. Ecol Monogr 35:1–35
Stiller J, da Fonseca RR, Alfaro ME, Faircloth BC, Wilson NG, Rouse GW (2021) Using ultraconserved elements to track the influence of sea-level change on leafy seadragon populations. Mol Ecol 30:1364–1380
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tang K, Stiassny MLJ, Mayden RL, DeSalle R (2021) Systematics of Damselfishes. Ichthyol Herpetol 109:258–318
Tavera J, Arturo AP, Wainwright PC (2018) Multilocus phylogeny, divergence times, and a major role for the benthic-to-pelagic axis in the diversification of grunts (Haemulidae). Mol Phylogenet Evol 121:212–223
Tavera J, Wainwright PC (2019) Geography of speciation affects rate of trait divergence in haemulid fishes. Proc Roy Soc B Biol Sci 286:20182852
Tea YK, Xu X, DiBattista JD, Lo N, Cowman PF, Ho SY (2021) Phylogenomic Analysis of Concatenated Ultraconserved Elements Reveals the Recent Evolutionary Radiation of the Fairy Wrasses (Teleostei: Labridae: Cirrhilabrus). Syst Biol: https://doi.org/10.1093/sysbio/syab012
Tittensor DP, Mora C, Jetz W, Lotze HK, Ricard D, Berghe EV, Worm B (2010) Global patterns and predictors of marine biodiversity across taxa. Nature 466:1098–1101
Venables WN, Ripley BD (2002) Modern Applied Statistics with S, Fourth edition. Springer, New York. ISBN 0–387–95457–0, https://www.stats.ox.ac.uk/pub/MASS4/
Victor BC (2015) How many coral reef fish species are there? Cryptic diversity and the new molecular taxonomy. In: Mora C (ed) Ecology of fishes on coral reefs the functioning of an ecosystem in a changing World. Cambridge University Press, Cambridge
Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD (2005) DNA barcoding Australia’s fish species. Phil Trans Roy Soc B Biol Sci 360:1847–1857
Webb PW, LaLiberte GD, Schrank AJ (1996) Does body and fin form affect the maneuverability of fish traversing vertical and horizontal slits? Environ Biol Fishes 46:7–14
Wilson S, Bellwood DR (1997) Cryptic dietary components of territorial damselfishes (Pomacentridae, Labroidei). Mar Ecol Prog Ser 153:299–310
WoRMS Editorial Board (2021) World Register of Marine Species. Available from http://www.marinespecies.org at VLIZ. doi:https://doi.org/10.14284/170
Acknowledgements
This research was informed by the helpful advice of Malcolm Francis, Clinton Duffy and Gerry Allen. Expeditions to Rangitāhua, Rapa Iti, Pitcairn Islands and New Caledonia were made possible by the crew of the RV Braveheart, Stoney Creek Shipping Company. For field assistance to collect specimens in New Caledonia we thank Jeremy Barker, Carl Struthers, Irene Middleton, Mark McGrouther and Sally Reader; in Norfolk Island we thank Mark Scott; in Rangitāhua we thank Alison Ballance, Charles Bedford, Clinton Duffy, Malcolm Francis, Sam McCormack, Mark McGrouther, Phil Ross, Carl Struthers, Stephen Ullrich, Ged Wiren and Vincent Zintzen; in Rapa Iti we thank Charles Bedford, Anna Berthelsen, Amelia Connell, Mark McGrouther, Sally Reader, Carina Sim-Smith and Stephen Ullrich. We wish to thank and acknowledge mana whenua (the traditional owners) of Rangitāhua, the Māori iwi of Ngāti Kuri for their support and the New Zealand Department of Conservation for sampling permission (Authorisation number: 47976 MAR). These expeditions were supported by Natural History New Zealand and the New Zealand on Air Platinum Fund, the Auckland Museum Institute, Auckland Museum, the Pew Charitable Trusts and the School of Natural and Computational Sciences Massey University. In support of fieldwork in Rapa Nui, we thank Cristian Rapu, Valentina Neglia, Emily C. Giles, but also Rebeca Tepano, Nina, Taveke Olivares Rapu, Liza Garrido Toleado from SERNAPESCA and Ludovic Burns Tuki from Mesa del Mar, and the people of the Rapa Nui Island for their kind and generous support. Specimens were collected under permit No. 724, 8 March 2016 obtained from the Chilean Subsecretary of Fishing. All applicable institutional guidelines for the care and use of animals were followed. The Universidad Austral de Chile Ethical Care Committee and Biosecurity Protocol approved our use and handling of animals. Specimens in French Polynesia were collected under annual research permits (unnumbered) issued by the French Polynesian Ministry of Research to the CRIOBE. We are grateful for the specimens and tissues provided by Mark McGrouther at the Australian Museum; David Catania at the California Academy of Sciences; and Arnold Suzumoto at the Bernice P. Bishop Museum. We are grateful to Amy Driskell and Ross Robertson of the Smithsonian Institution for allowing access to unpublished C. rapanui sequences from Rapa Nui that motivated this study. Images were supplied by Ian Skipworth, Irene Middleton (seacologynz.com) and Luiz Rocha, California Academy of Sciences. LL and JDA were supported by Rutherford Foundation New Zealand Postdoctoral fellowships, and EDT was supported by a Fondecyt Postdoctorado Fellowship n°3160692 (Chile). Ongoing biodiversity research of Rangitāhua is supported by a New Zealand Ministry of Business, Innovation, and Employment Endeavour Research Fund “Te Mana o Rangitāhua” (69495-ENDRP-AWMMU2001) and Rutherford Discovery Fellowships to LL (RDF-20-MAU-001) and JDA (RDF-19-MAU-006). Comments from two reviewers and the editor greatly improved an earlier version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional guidelines for the care and use of animals were followed. All specimens were collected, and all research carried out, with appropriate national permissions. All raw sequence data and sample metadata are deposited in appropriate open-access repositories (raw sequences at INSDC, Accession numbers: KM455353-KM455364, MK657005, MK657007, MK657116, MK657705, MK657836, MK658695, MZ832606-MZ832613, MZ852575-MZ852644; sample metadata at GEOME [https://n2t.net/ark:/21547/Dvj2] as part of the Ira Moana Project, https://sites.massey.ac.nz/iramoana/). Some of these samples and derived data have a Biocultural (BC) Notice attached. The BC Notice is a visible notification that there are accompanying cultural rights and responsibilities that need further attention for any future sharing and use of this material or data. The BC Notice recognises the rights of Indigenous peoples to permit the use of information, collections, data and digital sequence information generated from the biodiversity or genetic resources associated with traditional lands, waters and territories. The BC Notice may indicate that BC (Biocultural) Labels are in development and their implementation is being negotiated. For more information about the BC Notices, visit https://localcontexts.org/notice/bc-notice/. All authors contributed to the generation of the research: conceptual development (LL, TT, EDT, PSA, SP, JDA, LK), funding (LL, TT, PSA, SP), field research (LL, TT, EDT, PSA, SP, JDA), laboratory analysis (EDT, LL, VA, JIC, LK, TT), statistical analysis (JDA, LL, TT, EDT, JIC, LK), writing and editing (LL, JDA, EDT, TT, PSA, LK, JIC). All funding sources are declared in the Acknowledgements section.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Toplic Editor Morgan S. Pratchett
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liggins, L., Kilduff, L., Trnski, T. et al. Morphological and genetic divergence supports peripheral endemism and a recent evolutionary history of Chrysiptera demoiselles in the subtropical South Pacific. Coral Reefs 41, 797–812 (2022). https://doi.org/10.1007/s00338-021-02179-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-021-02179-7