Abstract
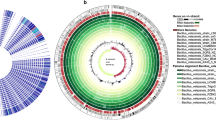
The application of biocontrol biopesticides based on plant growth–promoting rhizobacteria (PGPR), particularly members of the genus Bacillus, is considered a promising perspective to make agricultural practices sustainable and ecologically safe. Recent advances in genome sequencing by third-generation sequencing technologies, e.g., Pacific Biosciences’ Single Molecule Real-Time (PacBio SMRT) platform, have allowed researchers to gain deeper insights into the molecular and genetic mechanisms of PGPR activities, and to compare whole genome sequences and global patterns of epigenetic modifications. In the current work, this approach was used to sequence and compare four Bacillus strains that exhibited various PGPR activities including the strain UCMB5140, which is used in the commercial biopesticide Phytosubtil. Whole genome comparison and phylogenomic inference assigned the strain UCMB5140 to the species Bacillus velezensis. Strong biocontrol activities of this strain were confirmed in several bioassays. Several factors that affect the evolution of active PGPR B. velezensis strains were identified: (1) horizontal acquisition of novel non-ribosomal peptide synthetases (NRPS) and adhesion genes; (2) rearrangements of functional modules of NRPS genes leading to strain specific combinations of their encoded products; (3) gain and loss of methyltransferases that can cause global alterations in DNA methylation patterns, which eventually may affect gene expression and regulate transcription. Notably, we identified a horizontally transferred NRPS operon encoding an uncharacterized polypeptide antibiotic in B. velezensis UCMB5140. Other horizontally acquired genes comprised a possible adhesin and a methyltransferase, which may explain the strain-specific methylation pattern of the chromosomal DNA of UCMB5140.
Key points
• Whole genome sequence of the active PGPR Bacillus velezensis UCMB5140.
• Identification of genetic determinants responsible for PGPR activities.
• Role of methyltransferases and epigenetic mechanisms in evolution of bacteria.






Similar content being viewed by others
References
Abd El-Daim IA, Bejai S, Meijer J (2019) Bacillus velezensis 5113 induced metabolic and molecular reprogramming during abiotic stress tolerance in wheat. Sci Rep 9:16282. https://doi.org/10.1038/s41598-019-52567-x
Aloo BN, Makumba BA, Mbega ER (2019) The potential of bacilli rhizobacteria for sustainable crop production and environmental sustainability. Microbiol Res 219:26–39. https://doi.org/10.1016/j.micres.2018.10.011
Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA (2016) plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. https://doi.org/10.1093/bioinformatics/btw493
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F (2008) The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. https://doi.org/10.1186/1471-2164-9-75
Babasaki K, Takao T, Shimonishi Y, Kurahashi K (1985) Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J Biochem 98:585–603. https://doi.org/10.1093/oxfordjournals.jbchem.a135315
Beaulaurier J, Schadt EE, Fang G (2019) Deciphering bacterial epigenomes using modern sequencing technologies. Nat Rev Genet 20:157–172. https://doi.org/10.1038/s41576-018-0081-3
Bezuidt O, Lima-Mendez G, Reva ON (2009) SEQWord Gene Island sniffer: a program to study the lateral genetic exchange among bacteria. World Acad Sci Eng Technol 58:1169–1174
Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T (2019) antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. https://doi.org/10.1093/nar/gkz310
Caulier S, Nannan C, Gillis A, Licciardi F, Bragard C, Mahillon J (2019) Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Front Microbiol 10:302. https://doi.org/10.3389/fmicb.2019.00302
Chaisson MJ, Tesler G (2012) Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13:238. https://doi.org/10.1186/1471-2105-13-238
Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R (2006) Structural and functional characterization of three polyketide synthase gene clusters in Bacillus amyloliquefaciens FZB 42. J Bacteriol 188:4024–4036. https://doi.org/10.1128/JB.00052-06
Chen XH, Koumoutsi A, Scholz R, Schneider K, Vater J, Süssmuth R, Piel J, Borriss R (2009a) Genome analysis of Bacillus amyloliquefaciens FZB42 reveals its potential for biocontrol of plant pathogens. J Biotechnol 140:27–37. https://doi.org/10.1016/j.jbiotec.2008.10.011
Chen XH, Scholz R, Borriss M, Junge H, Mögel G, Kunz S, Borriss R (2009b) Difficidin and bacilysin produced by plant associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J Biotechnol 140:38–44. https://doi.org/10.1016/j.jbiotec.2008.10.015
Darling AE, Mau B, Perna NT (2010) progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. https://doi.org/10.1371/journal.pone.0011147
De Vrieze M, Varadarajan AR, Schneeberger K, Bailly A, Rohr RP, Ahrens CH, Weisskopf L (2020) Linking comparative genomics of nine potato-associated Pseudomonas isolates with their differing biocontrol potential against late blight. Front Microbiol 11:857. https://doi.org/10.3389/fmicb.2020.00857
Delsuc F, Brinkmann H, Philippe H (2005) Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet 6:361–375. https://doi.org/10.1038/nrg1603
Dunlap CA, Bowman MJ, Rooney AP (2019) Iturinic lipopeptide diversity in the Bacillus subtilis species group—important antifungals for plant disease biocontrol applications. Front Microbiol 10:1794. https://doi.org/10.3389/fmicb.2019.01794
Emms DM, Kelly S (2015) OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16:157. https://doi.org/10.1186/s13059-015-0721-2
Fan B, Wang C, Ding X, Zhu B, Song X, Borriss R (2019, 2019) AmyloWiki: an integrated database for Bacillus velezensis FZB42, the model strain for plant growth-promoting bacilli. Database (Oxford):baz071. https://doi.org/10.1093/database/baz071
Ferrarini M, Moretto M, Ward JA, Šurbanovski N, Stevanović V, Giongo L, Viola R, Cavalieri D, Velasco R, Cestaro A, Sargent DJ (2013) An evaluation of the PacBio RS platform for sequencing and de novo assembly of a chloroplast genome. BMC Genomics 14:670. https://doi.org/10.1186/1471-2164-14-670
Freeman JM, Plasterer TN, Smith TF, Mohr SC (1998) Patterns of genome organization in bacteria. Science 279:1827–11827. https://doi.org/10.1126/science.279.5358.1827a
Garrison E, Marth G (2012) Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907
Genin S, Denny TP (2012) Pathogenomics of the Ralstonia solanacearum species complex. Annu Rev Phytopathol 50:67–89. https://doi.org/10.1146/annurev-phyto-081211-173000
Gong AD, Li HP, Yuan QS, Song XS, Yao W, He WJ, Zhang JB, Liao YC (2015) Antagonistic mechanism of iturin A and plipastatin A from Bacillus amyloliquefaciens S76-3 from wheat spikes against Fusarium graminearum. PLoS One 10:e0116871. https://doi.org/10.1371/journal.pone.0116871
Gu Q, Yang Y, Yuan Q, Shi G, Wu L, Lou Z, Huo R, Wu H, Borriss R, Gao X (2017) Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant pathogenic fungus Fusarium graminearum. Appl Environ Microbiol 83:e01075–e01017. https://doi.org/10.1128/AEM.01075-17
Guo W, Cui P, Chen X (2015) Complete genome of Bacillus sp. Pc3 isolated from the Antarctic seawater with antimicrobial activity. Mar Genomics 20:1–2. https://doi.org/10.1016/j.margen.2015.01.004
Gupta CD, Dubey RC, Kang SC, Maheshwari DK (2001) Antibiosis-mediated necrotrophic effect of Pseudomonas GRC2 against two fungal plant pathogens. Curr Sci 81:91–94
Hashem A, Tabassum B, Fathi Abd Allah E (2019) Bacillus subtilis: a plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J Biol Sci 26:1291–1297. https://doi.org/10.1016/j.sjbs.2019.05.004
Idriss EE, Makarewicz O, Farouk A, Rosner K, Greiner R, Bochow H, Richter T, Borriss R (2002) Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiol 148(Pt 7):2097–2109. https://doi.org/10.1099/00221287-148-7-2097
Iyer S, Acharya KR (2011) Tying the knot: the cystine signature and molecular-recognition processes of the vascular endothelial growth factor family of angiogenic cytokines. FEBS J 278:4304–4322. https://doi.org/10.1111/j.1742-4658.2011.08350.x
Khan N, Bano A, Rahman MA, Guo J, Kang Z, Babar MA (2019) Comparative physiological and metabolic analysis reveals a complex mechanism involved in drought tolerance in chickpea (Cicer arietinum L.) induced by PGPR and PGRs. Sci Rep 9:2097. https://doi.org/10.1038/s41598-019-38702-8
Khokhar MK, Tetarwal JP (2012) Management of post-harvest black mould fruit rot of pomegranate (Punica granatum L.) caused by Aspergillus niger (Tieghem). Agric Res Rev 1:162–165
Kim YH, Choi Y, Oh YY, Ha NC, Song J (2019) Plant growth-promoting activity of beta-propeller protein YxaL secreted from Bacillus velezensis strain GH1-13. PLoS One 14:e0207968. https://doi.org/10.1371/journal.pone.0207968
Kolmogorov M, Yuan J, Lin Y, Pevzner PA (2019) Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. https://doi.org/10.1038/s41587-019-0072-8
Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J Bacteriol 186:1084–1096. https://doi.org/10.1128/jb.186.4.1084-1096.2004
Landy M, Warren GH, Roseman SB, Colio LG (1948) Bacillomycin, an antibiotic from Bacillus subtilis active against pathogenic fungi. Proc Soc Exp Biol Med 67:539–541. https://doi.org/10.3181/00379727-67-16367
Li Y, Gu Y, Li J, Xu M, Wei Q, Wang Y (2015) Biocontrol agent Bacillus amyloliquefaciens LJ02 induces systemic resistance against cucurbits powdery mildew. Front Microbiol 6:1–15. https://doi.org/10.3389/fmicb.2015.00883
Liu Z, Budiharjo A, Wang P, Shi H, Fang J, Borriss R, Zhang K, Huang X (2013) The highly modified microcin peptide plantazolicin is associated with nematicidal activity of Bacillus amyloliquefaciens FZB42. Appl Microbiol Biotechnol 97:10081–10090. https://doi.org/10.1007/s00253-013-5247-5
Lu X, Liu SF, Yue L, Zhao X, Zhang YB, Xie ZK, Wang RY (2018) Epsc involved in the encoding of exopolysaccharides produced by Bacillus amyloliquefaciens FZB42 act to boost the drought tolerance of Arabidopsis thaliana. Int J Mol Sci 19:E3795. https://doi.org/10.3390/ijms19123795
Manzoor S, Niazi A, Bejai S, Meijer J, Bongcam-Rudloff E (2013) Genome sequence of a plant-associated bacterium, Bacillus amyloliquefaciens strain UCMB5036. Genome Announc 1:e0011113. https://doi.org/10.1128/genomeA.00111-13
Marczynski GT, Shapiro L (1993) Bacterial chromosome origins of replication. Curr Opin Genet Dev 3:775–782. https://doi.org/10.1016/s0959-437x(05)80098-x
Martinez-Garcia E, Fraile S, Rodriguez-Espeso D, Vecchietti D, Bertoni G, de Lorenzo V (2020) The naked cell: emerging properties of a surfome-streamlined Pseudomonas putida strain. bioRxiv. https://doi.org/10.1101/2020.05.17.100628
McErlean M, Overbay J, Van Lanen S (2019) Refining and expanding nonribosomal peptide synthetase function and mechanism. J Ind Microbiol Biotechnol 46:493–513. https://doi.org/10.1007/s10295-018-02130-w
Narasimhan V, Danecek P, Scally A, Xue Y, Tyler-Smith C, Durbin R (2016) BCFtools/RoH: a hidden Markov model approach for detecting autozygosity from next-generation sequencing data. Bioinformatics 32:1749–1751. https://doi.org/10.1093/bioinformatics/btw044
Niazi A, Manzoor S, Asari S, Bejai S, Meijer J, Bongcam-Rudloff E (2014) Genome analysis of Bacillus amyloliquefaciens subsp. plantarum UCMB5113: a rhizobacterium that improves plant growth and stress management. PLoS One 9:e104651. https://doi.org/10.1371/journal.pone.0104651
Omasits U, Varadarajan AR, Schmid M, Goetze S, Melidis D, Bourqui M, Nikolayeva O, Québatte M, Patrignani A, Dehio C, Frey JE, Robinson MD, Wollscheid B, Ahrens CH (2017) An integrative strategy to identify the entire protein coding potential of prokaryotic genomes by proteogenomics. Genome Res 27:2083–2095. https://doi.org/10.1101/gr.218255.116
Otusanya MO, Jeger MJ (1996) Effect of Aspergillus niger on shoot emergence and vine development in field-sown yams (Dioscorea spp.) and rot development under long-term storage conditions. Int Biodeterior Biodegrad 38:89–100. https://doi.org/10.1016/S0964-8305(96)00030-3
Pierneef R, Cronje L, Bezuidt O, Reva ON (2015) Pre_GI: a global map of ontological links between horizontally transferred genomic islands in bacterial and archaeal genomes. Database (Oxford) 2015:bav058. https://doi.org/10.1093/database/bav058
Pushpanathan M, Gunasekaran P, Rajendhran J (2013) Antimicrobial peptides: versatile biological properties. Int J Pept 2013:675391–675315. https://doi.org/10.1155/2013/675391
Qin M, Wu S, Li A, Zhao F, Feng H, Ding L, Ruan J (2019) LRScaf: improving draft genomes using long noisy reads. BMC Genomics 20:955. https://doi.org/10.1186/s12864-019-6337-2
Rabbee MF, Ali MS, Choi J, Hwang BS, Jeong SC, Baek KH (2019) Bacillus velezensis: a valuable member of bioactive molecules within plant microbiomes. Molecules 24:E1046. https://doi.org/10.3390/molecules24061046
Raza W, Ling N, Yang L, Huang Q, Shen Q (2016) Response of tomato wilt pathogen Ralstonia solanacearum to the volatile organic compounds produced by a biocontrol strain Bacillus amyloliquefaciens SQR-9. Sci Rep 6:1–13. https://doi.org/10.1038/srep24856
Reva ON, Swanevelder DZH, Mwita LA, Mwakilili AD, Muzondiwa D, Joubert M, Chan WY, Lutz S, Ahrens CH, Avdeeva LV, Kharkhota MA, Tibuhwa D, Lyantagaye S, Vater J, Borriss R, Meijer J (2019) Genetic, epigenetic and phenotypic diversity of four Bacillus velezensis strains used for plant protection or as probiotics. Front Microbiol 10:2610. https://doi.org/10.3389/fmicb.2019.02610
Rhoads A, Au KF (2015) PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13:278–289. https://doi.org/10.1016/j.gpb.2015.08.002
Ryu C-M, Farag MA, Hu C-H, Reddy MS, Kloepper JW, Pare PW (2004) Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol 134:1017–1026. https://doi.org/10.1104/pp.103.026583
Safronova LA, Zhmurko LG, Karlovsky OA, Evseenko OV (2012) New environmentally safe biopreparation as an effective means of protection against soybean disease. Agroecol J 4:71–76
Safronova LA, Nudga AY, Avdeeva LV (2013) Patent of Ukraine №. 77141. Phytosubtil biopreparation for processing root crops and potatoes during their storage. Bul. № 2, 01/25/2013
Sánchez-Romero MA, Casadesús J (2020) The bacterial epigenome. Nat Rev Microbiol 18:7–20. https://doi.org/10.1038/s41579-019-0286-2
Saxena AK, Kumar M, Chakdar H, Anuroopa N, Bagyaraj DJ (2019) Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol 128:1583–1594. https://doi.org/10.1111/jam.14506
Schmid M, Frei D, Patrignani A, Schlapbach R, Frey JE, Remus-Emsermann MNP, Ahrens CH (2018) Pushing the limits of de novo genome assembly for complex prokaryotic genomes harboring very long, near identical repeats. Nucleic Acids Res 46:8953–8965. https://doi.org/10.1093/nar/gky726
Schneider K, Chen XH, Vater J, Franke P, Nicholson G, Borriss R, Süssmuth RD (2007) Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J Nat Prod 70:417–1423. https://doi.org/10.1021/np070070k
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. https://doi.org/10.1093/bioinformatics/btv351
Sović I, Šikić M, Wilm A, Fenlon SN, Chen S, Nagarajan N (2016) Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun 7:11307. https://doi.org/10.1038/ncomms11307
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Teng JLL, Yeung ML, Chan E, Jia L, Lin CH, Huang Y, Tse H, Wong SSY, Sham PC, Lau SKP, Woo PCY (2017) PacBio but not Illumina technology can achieve fast, accurate and complete closure of the high GC, complex Burkholderia pseudomallei two-chromosome genome. Front Microbiol 8:1448. https://doi.org/10.3389/fmicb.2017.01448
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14:178–192. https://doi.org/10.1093/bib/bbs017
Varadarajan AR, Allan RN, Valentin JD, Ocampo OEC, Somerville V, Pietsch F, Buhmann MT, West JJ, Skipp PJ, van der Mei HC, Ren Q, Schreiber F, Webb JS, Ahrens, CH. (2020) An integrated model system to gain mechanistic insights into biofilm formation and antimicrobial resistance development in Pseudomonas aeruginosa MPAO1. bioRxiv. https://doi.org/10.1101/2020.02.06.936690
Vaser R, Sović I, Nagarajan N, Šikić M (2017) Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27:737–746
Vater J, Kablitz B, Wilde C, Franke P, Mehta N, Cameotra SS (2002) Matrix-assisted laser desorption ionization–time of flight mass spectrometry of lipopeptide biosurfactants in whole cells and culture filtrates of Bacillus subtilis C-1 isolated from petroleum sludge. Appl Environ Microbiol 68:6210–6219. https://doi.org/10.1128/aem.68.12.6210-6219.2002
Wu LM, Wu HJ, Chen LN, Xie S, Zang H, Borriss R, Gao X (2014) Bacilysin from Bacillus amyloliquefaciens FZB42 has specific bactericidal activity against harmful algal bloom species. Appl Environ Microbiol 80:7512–7520. https://doi.org/10.1128/AEM.02605-14
Wu LM, Wu HJ, Chen L, Yu X, Borriss R, Gao X (2015) Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci Rep 5:12975. https://doi.org/10.1038/srep12975
Wu L, Huang Z, Li X, Ma L, Gu Q, Wu H, Liu J, Borriss R, Wu Z, Gao X (2018) Stomatal closure and SA-, JA/ET-signaling pathways are essential for Bacillus amyloliquefaciens FZB42 to restrict leaf disease caused by Phytophthora nicotianae in Nicotiana benthamiana. Front Microbiol 9:847. https://doi.org/10.3389/fmicb.2018.00847
Yuan J, Raza W, Shen Q, Huang Q (2012) Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl Environ Microbiol 78:5942–5944. https://doi.org/10.1128/AEM.01357-12
Zhao J, Yuan S, Gao B, Zhu S (2018) Molecular diversity of fungal inhibitor cystine knot peptides evolved by domain repeat and fusion. FEMS Microbiol Lett 365:365. https://doi.org/10.1093/femsle/fny158
Zhu B, Stülke J (2017) SubtiWiki in 2018: from genes and proteins to functional network annotation of the model organism Bacillus subtilis. Nucleic Acids Res 46:D743–D748. https://doi.org/10.1093/nar/gkx908
Acknowledgments
We gratefully acknowledge K. Dietel (ABiTEP GmbH, Berlin, Germany) for preparing samples for mass spectroscopic measurements, and Daniel Frei and Juerg E. Frey (Agroscope, Switzerland) for the Illumina MiSeq sequencing.
Funding
O.N.R. was funded by the South African National Research Foundation (NRF) grants 93134 and 93664; O.N.R., Sy.L., and D.T. received funds from the joint NRF/COSTECH (Tanzanian Commission for Science and Technology) grant 86905 and from joint TIA (Technology Innovation Agency of South Africa)/COSTECH grant TIA 2018-FUN-0050. A.D.M. acquired PhD and MSc student fellowship grants from Southern African Biochemistry and Informatics for Natural Products (SABINA, http://www.sabina-africa.org/). C.H.A. acknowledges support from Agroscope through its research program Microbial Biodiversity.
Author information
Authors and Affiliations
Contributions
O.N.R.—DNA and RNA sequencing, genome assembly, annotation, epigenetic profiling, funding acquisition, student supervision; S.A.L.—UCMB5140 isolation and characterization; A.D.M.—PGPR assays; D.M.—PGPR assays, student supervision; Sy.L.—PGPR assays, student supervision; W.Y.C.—mass spectrometry studies; St.L.—PacBio and Illumina MiSeq sequencing, genome assembly; C.H.A.—funding acquisition, PacBio and Illumina MiSeq sequencing, genome assembly; J.V.—mass spectroscopic measurements; R.B.—mass spectrometry and study supervision. All co-authors took part in the manuscript writing and edition.
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies with human participants or animals performed by any of the authors. All genome sequences obtained during the present study were made publicly accessible from NCBI database.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 3774 kb)
Rights and permissions
About this article
Cite this article
Reva, O.N., Larisa, S.A., Mwakilili, A.D. et al. Complete genome sequence and epigenetic profile of Bacillus velezensis UCMB5140 used for plant and crop protection in comparison with other plant-associated Bacillus strains. Appl Microbiol Biotechnol 104, 7643–7656 (2020). https://doi.org/10.1007/s00253-020-10767-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00253-020-10767-w