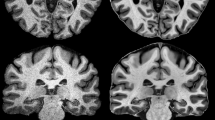
Abstract
Segmentation of human brain using structural MRI is a key step of processing in imaging neuroscience. The methods have undergone a rapid development in the past two decades and are now widely available. This non-technical review aims at providing an overview and basic understanding of the most common software. Starting with the basis of structural MRI contrast in brain and imaging protocols, the concepts of voxel-based and surface-based segmentation are discussed. Special emphasis is given to the typical contrast features and morphological constraints of cortical and sub-cortical grey matter. In addition to the use for voxel-based morphometry, basic applications in quantitative MRI, cortical thickness estimations, and atrophy measurements as well as assignment of cortical regions and deep brain nuclei are briefly discussed. Finally, some fields for clinical applications are given.








Similar content being viewed by others
References
Lim KO, Pfefferbaum A (1989) Segmentation of MR brain images into cerebrospinal fluid spaces, white and gray matter. J Comput Assist Tomogr 13:588–593
Ashburner J (2012) SPM: a history. Neuroimage 62:791–800
Lucas B, Bogovic J, Carass A, Bazin P-L, Prince J, Pham D, Landman B (2010) The Java image science toolkit (JIST) for rapid prototyping and publishing of neuroimaging software. Neuroinformatics 18:5–17
Fedorov A, Beichel R, Kalpathy-Cramer J, Finet J, Fillion-Robin J-C, Pujol S, Bauer C, Jennings D, Fennessy F, Sonka M, Buatti J, Aylward SR, Miller JV, Pieper S, Kikinis R (2012) 3D Slicer as an image computing platform for the quantitative imaging network. Magn Reson Imaging 30(9):1323–1341
Warfield SK, Zou KH, Wells WM (2004) Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Trans Med Imaging 23(7):903–921
Koenig SH, Brown RD 3rd, Spiller M, Lundbom N (1990) Relaxometry of brain: why white matter appears bright in MRI. Magn Reson Med 14(3):482–495
Kamman RL, Go KG, Brouwer W, Berendsen HJ (1988) Nuclear magnetic resonance relaxation in experimental brain edema: effects of water concentration, protein concentration, and temperature. Magn Reson Med 6(3):265–274
Gelman N, Gorell JM, Barker PB, Savage RM, Spickler EM, Windham JP, Knight RA (1999) MR imaging of human brain at 3.0 T: preliminary report on transverse relaxation rates and relation to estimated iron content. Radiology 210(3):759–767
Gelman N, Ewing JR, Gorell JM, Spickler EM, Solomon EG (2001) Interregional variation of longitudinal relaxation rates in human brain at 3.0 T: relation to estimated iron and water contents. Magn Reson Med 45(1):71–79
Hallgren B, Sourander P (1958) The effect of age on the non-haemin iron in the human brain. J Neurochem 3(1):41–51
Helms G, Kallenberg K, Dechent P (2006) A contrast-driven approach to intracranial segmentation using a combination of T2- and T1-weighted 3D MRI datasets. J Magn Reson Imaging 24(4):790–795
Deichmann R, Good CD, Josephs O, Ashburner J, Turner R (2000) Optimization of 3-D MP-RAGE sequences for structural brain imaging. Neuroimage 12:112–127
Deichmann R, Schwarzbauer C, Turner R (2004) Optimisation of the 3D MDEFT sequence for anatomical brain imaging: technical implications at 1.5 and 3 T. NeuroImage 21:757–767
Jack CJ, Bernstein M, Fox N, Thompson P, Alexander G, Harvey D, Borowski B, Britson P, Whitwell J, Ward C, Dale A, Felmlee J, Gunter J, Hill D, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli C, Krueger G, Ward H, Metzger G, Scott K, Mallozzi R, Blezek D, Levy J, Debbins J, Fleisher A, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner M (2008) The Alzheimer’s disease neuroimaging initiative (ADNI): MRI methods. J Magn Reson Imaging 27(4):685–691
Alfano B, Brunetti A, Covelli EM, Quarantelli M, Panico MR, Ciarmiello A, Salvatore M (1997) Unsupervised, automated segmentation of the normal brain using a multispectral relaxometric magnetic resonance approach. Magn Reson Med 37(1):84–93
Sled JG, Zijdenbos AP, Evans AC (1998) A non-parametric method for automatic correction of intensity non-uniformity in MRI. IEEE Trans Med Imag 17:87–97
Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imag 29(6):1310–1320
Zhang Y, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation–maximization algorithm. IEEE Trans Med Imaging 20(1):45–57
Smith SM (2002) Fast robust automated brain extraction. Hum Brain Mapp 17(3):143–155
Iglesias JE, Liu CY, Thompson PM, Tu Z (2011) Robust brain extraction across datasets and comparison with publicly available methods. IEEE Trans Med Imaging 30:1617–1634
Ségonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, Fischl B (2004) A hybrid approach to the skull stripping problem in MRI. Neuroimage 22(3):1060–1075
Carass A, Cuzzocreo J, Wheeler MB, Bazin PL, Resnick SM, Prince JL (2011) Simple paradigm for extra-cerebral tissue removal: algorithm and analysis. Neuroimage 56(4):1982–1992
Mikheev A, Nevsky G, Govindan S, Grossman R, Rusinek HJ (2008) Fully automatic segmentation of the brain from T1-weighted MRI using Bridge Burner algorithm. J Magn Reson Imaging 27(6):1235–1241
Woolrich MW, Jbabdi S, Patenaude B, Chappell M, Makni S, Behrens T, Beckmann C, Jenkinson M, Smith SM (2009) Bayesian analysis of neuroimaging data in FSL. Neuroimage 45:S173–S186
Ashburner J, Friston K (1997) Multimodal image coregistration and partitioning—a unified framework. Neuroimage 6(3):209–217
Ashburner J, Friston KJ (2005) Unified segmentation. Neuroimage 26(3):839–851
Evans AC, Kamber M, Collins DL, Macdonald D (1994) An MRI-based probabilistic atlas of neuroanatomy. In: Shorvon S, Fish D, Andermann F, Bydder GM, Stefan H (eds) Magnetic resonance scanning and epilepsy, NATO ASI series A, life sciences, vol 264. Plenum, New York, pp 263–274
van Leemput K, Maes F, Vandermeulen D, Suetens P (1999) Automated model-based tissue classification of MR images of the brain. IEEE Trans Med Imaging 18(10):897–908
Draganski B, Gaser C, Busch V, Schuierer G, Bogdahn U, May A (2004) Neuroplasticity: changes in grey matter induced by training. Nature 427(6972):311–312
Focke NK, Helms G, Kaspar S, Diederich C, Tóth V, Dechent P, Mohr A, Paulus W (2011) Multi-site voxel-based morphometry—not quite there yet. Neuroimage 56(3):1164–1170
Lambert C, Lutti A, Helms G, Frackowiak R, Ashburner J (2013) Multiparametric brainstem segmentation using a modified multivariate mixture of gaussians. Neuroimage Clin 16(2):684–694
Bazin P-L, Pham D (2007) Topology correction of segmented medical images using a fast marching algorithm. Comput Methods Programs Biomed 88:182–190
Bazin P-L, Pham D (2008) Homeomorphic brain image segmentation with topological and statistical atlases. Med Image Anal 12:616–625
Brownstein KR, Tarr CE (1977) Spin-lattice relaxation in a system governed by diffusion. J Magn Reson 26:17–24
Fischl B, Dale AM (2000) Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA 97(20):11050–11055
Kim JS, Singh V, Lee JK, Lerch J, Ad-Dab’bagh Y, MacDonald D, Lee JM, Kim SI, Evans AC (2005) Automated 3-D extraction and evaluation of the inner and outer cortical surfaces using a Laplacian map and partial volume effect classification. Neuroimage 27(1):210–221
Han X, Pham D, Tosun D, Rettmann M, Xu C, Prince J (2004) CRUISE: cortical reconstruction using implicit surface evolution. Neuroimage 23:997–1012
Dale AM, Fischl B, Sereno MI (1999) Cortical surface-based analysis I. Segmentation and surface reconstruction. Neuroimage 9:179–194
Kriegeskorte N, Goebel R (2001) An efficient algorithm for topologically correct segmentation of the cortical sheet in anatomical MR volumes. NeuroImage 14:329–346
Hutton C, De Vita E, Ashburner J, Deichmann R, Turner R (2008) Voxel-based cortical thickness measurements in MRI. Neuroimage 40(4):1701–1710
Hutton C, Draganski B, Ashburner J, Weiskopf N (2009) A comparison between voxel-based cortical thickness and voxel-based morphometry in normal aging. Neuroimage 48(2):371–380
Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RSR, Busa E, Morris JC, Dale AM, Fischl B (2004) Thinning of the cerebral cortex in aging. Cereb Cortex 14(7):721–730
Vachet C, Hazlett HC, Niethammer M, Oguz I, Cates J, Whitaker R, Piven J, Styner M (2011) Group-wise automatic mesh-based analysis of cortical thickness. In: Presented at the medical imaging 2011: image processing 7962(1):796227
Shi F, Yap P-T, Wu G, Jia H, Gilmore JH, Lin W, Shen D (2011) Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One 6(4):e18746
Jia H, Yap PT, Shen D (2012) Iterative multi-atlas-based multi-image segmentation with tree-based registration. Neuroimage 59(1):422–430
Wu G, Wang Q, Zhang D, Nie F, Huang H, Shen D (2014) A generative probability model of joint label fusion for multi-atlas based brain segmentation. Med Image Anal 18(6):881–890
Iglesias JE, Sabuncu MR (2015) Multi-atlas segmentation of biomedical images: a survey. Med Imag Anal 24(1):205–219
Cabezas M, Oliver A, Lladó X, Freixenet J, Cuadra MB (2011) A review of atlas-based segmentation for magnetic resonance brain images. Comput Methods Programs Biomed 104(3):e158–e177. doi:10.1016/j.cmpb.2011.07.015
Yushkevich PA, Pluta J, Wang H, Ding SL, Xie L, Gertje E, Mancuso L, Kliot D, Das SR, Wolk DA (2014) Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp 36(1):258–287
Yang Z, Y C, Bogovic JA, Carass A, Jedynak BM, Ying SH, Prince JL (2015) Automated cerebellar lobule segmentation with application to cerebellar structural analysis in cerebellar disease. Neuroimage doi:10.1016/j.neuroimage.2015.09.032. [Epub ahead of print]
Bogovic JA, Bazin PL, Ying SH, Prince JL (2013) Automated segmentation of the cerebellar lobules using boundary specific classification and evolution. Inf Process Med Imaging 23:62–73
Vachet C, Yvernault B, Bhatt K, Smith RG, Gerig G, Hazlett HC, Styner M (2012) Automatic corpus callosum segmentation using a deformable active Fourier contour model. Proc SPIE Int Soc Opt Eng 8317:831707. doi:10.1117/12.911504
Helms G, Draganski B, Frackowiak R, Ashburner J, Weiskopf N (2009) Reliable segmentation of deep brain grey matter structures using magnetization transfer (MT) parameter maps. Neuroimage 47:194–198
Deoni SC, Rutt BK, Parrent AG, Peters TM (2007) Segmentation of thalamic nuclei using a modified k-means clustering algorithm and high-resolution quantitative magnetic resonance imaging at 1.5 T. Neuroimage 34:117–126
Patenaude B, Smith SM, Kennedy D, Jenkinson M (2011) A Bayesian model of shape and appearance for subcortical brain. Neuroimage 56(3):907–922
Krauth A, Blanc R, Poveda A, Jeanmonod D, Morel A, Székely G (2010) A mean three-dimensional atlas of the human thalamus: generation from multiple histological data. Neuroimage 49(3):2053–2062
Hoult DI (2000) The principle of reciprocity in signal strength calculations—a mathematical guide. Concepts Magn Reson 14(4):173–187
Volz S, Nöth U, Deichmann R (2012) Correction of systematic errors in quantitative proton density mapping. Magn Reson Med 68(1):74–85
Weiskopf N, Lutti A, Helms G, Novak M, Ashburner J, Hutton C (2011) Unified segmentation based correction of R1 brain maps for RF transmit field inhomogeneities (UNICORT). Neuroimage 54(3):2116–2124
Shin W, Geng X, Gu H, Zhan W, Zou Q, Yang Y (2010) Automated brain tissue segmentation based on fractional signal mapping from inversion recovery look-locker acquisition. Neuroimage 52:1347–1354
Ahlgren A, Wirestam R, Ståhlberg F, Knutsson L (2014) Automatic brain segmentation using fractional signal modeling of a multiple flip angle, spoiled gradient-recalled echo acquisition. Magn Reson Mater Phy 27:551–565
Draganski B, Ashburner J, Hutton C, Kherif F, Frackowiak RS, Helms G, Weiskopf N (2011) Regional specificity of MRI contrast parameter changes in normal ageing revealed by voxel-based quantification (VBQ). Neuroimage 55(4):1423–1434
Càmara E, Bodammer N, Rodríguez-Fornells A, Tempelmann C (2007) Age-related water diffusion changes in human brain: a voxel-based approach. Neuroimage 34:1588–1599
Dick F, Tierney AT, Lutti A, Josephs O, Sereno MI, Weiskopf N (2012) In vivo functional and myeloarchitectonic mapping of human primary auditory areas. J Neurosci 32(46):16095–16105
Miller DH, Barkhof F, Frank JA, Parker GJM, Thompson AJ (2002) Measurement of atrophy in multiple sclerosis: pathological basis, methodological aspects and clinical relevance. Brain 125:1676–1692
Smith SM, Zhang Y, Jenkinson M, Chen J, Matthews PM, Federico A, De Stefano N (2002) Accurate, robust, and automated longitudinal and cross-sectional brain change analysis. Neuroimage 17:479–489
Durand-Dubief F, Belaroussi B, Armspach JP, Dufour M, Roggerone S, Vukusic S, Hannoun S, Sappey-Marinier D, Confavreux C, Cotton F (2012) Reliability of longitudinal brain volume loss measurements between 2 sites in patients with multiple sclerosis: comparison of 7 quantification techniques. AJNR Am J Neuroradiol 33(10):1918–1924
Bernasconi A, Bernasconi N, Bernhardt BC, Schrader D (2011) Advances in MRI for ‘cryptogenic’ epilepsies. Nat Rev Neurol 7(2):99–108
Martin P, Bender B, Focke NK (2015) Post-processing of structural MRI for individualized diagnostics. Quant Imaging Med Surg 5(2):188–203
Teipel SJ, Grothe M, Lista S, Toschi N, Garaci FG, Hampel H (2013) Relevance of magnetic resonance imaging for early detection and diagnosis of Alzheimer disease. Med Clin North Am 97(3):399–424
Braskie MN, Thompson PM (2014) A focus on structural brain imaging in the Alzheimer’s disease neuroimaging initiativ. Biol Psychiatry 75(7):527–533
Jack CR Jr, Barkhof F, Bernstein MA, Cantillon M, Cole PE, DeCarli C, Dubois B, Duchesne S, Fox NC, Frisoni GB, Hampel H, Hill DLG, Johnson K, Mangin J-F, Scheltens P, Schwarz AJ, Sperling R, Suhy J, Thompson PM, Weiner M, Foster NL (2011) Steps to standardization and validation of hippocampal volumetry as a biomarker in clinical trials and diagnostic criteria for Alzheimer’s disease. Alzheimers Dement 7(4):474–485
Horn A, Kühn AA (2014) Lead-DBS: a toolbox for deep brain stimulation electrode localizations and visualizations. Neuroimage 107:127–135
Shiee N, Bazin P-L, Ozturk A, Reich DS, Calabresi PA, Pham DL (2010) A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. Neuroimage 49:1524–1535
Prastawa M, Gerig G (2008) Brain Lesion Segmentation through physical model estimation. Int Symp Vis Comput (ISVC) Lect Notes Comput Sci (LNCS) 5358:562–571
Lao Z, Shen D, Liu D, Jawad AF, Melhem ER, Launer LJ, Bryan NR, Davatzikos C (2008) Computer-assisted segmentation of white matter lesions in 3D MR images using pattern recognition. Acad Radiol 15(3):300–313
Ithapu V, Singh V, Lindner C, Austin BP, Hinrichs C, Carlsson CM, Bendlin BB, Johnson SC (2014) Extracting and summarizing white matter hyperintensities using supervised segmentation methods in Alzheimer’s disease risk and aging studies. Hum Brain Mapp. doi:10.1002/hbm.22472
García-Lorenzo D, Francis S, Narayanan S, Arnold DL, Collins DL (2013) Review of automatic segmentation methods of multiple sclerosis white matter lesions on conventional magnetic resonance imaging. Med Image Anal 17(1):1–18
Moon N, Bullitt E, van Leemput K, Gerig G (2002) Automatic brain and tumor segmentation. In: Proceedings of MICCAI ‘02, Springer LNCS 2488, 09/2002
Bauer S, Wiest R, Nolte LP, Reyes M (2013) A survey of MRI-based medical image analysis for brain tumor studies. Phys Med Biol 58(13):R97–R129
Wang L, Shi F, Yap P-T, Lin W, Gilmore JH, Shen D (2013) Longitudinally guided level sets for consistent tissue segmentation of neonates. Hum Brain Mapp 34:956–972
Wang B, Prastawa M, Irimia A, Chambers MC, Sadeghi N, Vespa PM, van Horn JD, Gerig G (2013) Analyzing imaging biomarkers for traumatic brain injury using 4D modeling of longitudinal MRI. Proc IEEE Int Symp Biomed Imaging 2013:1392–1395
Shiee N, Bazin P-L, Zackowski KM, Farrell SK, Harrison DM, Newsome SD, Ratchford JN, Caffo BS, Calabresi PA, Pham DL, Reich DS (2012) Revisiting brain atrophy and its relationship to disability in multiple sclerosis. PLoS One 7(5):e37049. doi:10.1371/journal.pone.0037049
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
All procedures performed in studies involving human participants were accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Conflict of interest
The author has no conflicts of interest to declare.
Funding
The author is supported by the Swedish Research Council.
Rights and permissions
About this article
Cite this article
Helms, G. Segmentation of human brain using structural MRI. Magn Reson Mater Phy 29, 111–124 (2016). https://doi.org/10.1007/s10334-015-0518-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10334-015-0518-z