Abstract
Notch signalling plays a key role in the generation of haematopoietic stem cells (HSCs) during vertebrate development1,2,3 and requires intimate contact between signal-emitting and signal-receiving cells, although little is known regarding when, where and how these intercellular events occur. We previously reported that the somitic Notch ligands, Dlc and Dld, are essential for HSC specification4. It has remained unclear, however, how these somitic requirements are connected to the later emergence of HSCs from the dorsal aorta. Here we show in zebrafish that Notch signalling establishes HSC fate as their shared vascular precursors migrate across the ventral face of the somite and that junctional adhesion molecules (JAMs) mediate this required Notch signal transduction. HSC precursors express jam1a (also known as f11r) and migrate axially across the ventral somite, where Jam2a and the Notch ligands Dlc and Dld are expressed. Despite no alteration in the expression of Notch ligand or receptor genes, loss of function of jam1a led to loss of Notch signalling and loss of HSCs. Enforced activation of Notch in shared vascular precursors rescued HSCs in jam1a or jam2a deficient embryos. Together, these results indicate that Jam1a–Jam2a interactions facilitate the transduction of requisite Notch signals from the somite to the precursors of HSCs, and that these events occur well before formation of the dorsal aorta.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hadland, B. K. et al. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood 104, 3097–3105 (2004)
Yoon, M. J. et al. Mind bomb-1 is essential for intraembryonic hematopoiesis in the aortic endothelium and the subaortic patches. Mol. Cell. Biol. 28, 4794–4804 (2008)
Burns, C. E., Traver, D., Mayhall, E., Shepard, J. L. & Zon, L. I. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19, 2331–2342 (2005)
Clements, W. K. et al. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature 474, 220–224 (2011)
Weber, C., Fraemohs, L. & Dejana, E. The role of junctional adhesion molecules in vascular inflammation. Nature Rev. Immunol. 7, 467–477 (2007)
Sugano, Y. et al. Junctional adhesion molecule-A, JAM-A, is a novel cell-surface marker for long-term repopulating hematopoietic stem cells. Blood 111, 1167–1172 (2008)
Kobayashi, I. et al. Comparative gene expression analysis of zebrafish and mammals identifies common regulators in hematopoietic stem cells. Blood 115, e1–e9 (2010)
Thompson, M. A. et al. The cloche and spadetail genes differentially affect hematopoiesis and vasculogenesis. Dev. Biol. 197, 248–269 (1998)
Jin, S. W., Beis, D., Mitchell, T., Chen, J. N. & Stainier, D. Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development 132, 5199–5209 (2005)
Bertrand, J. Y. et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 464, 108–111 (2010)
Kissa, K. & Herbomel, P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 464, 112–115 (2010)
Qian, F. et al. Distinct functions for different scl isoforms in zebrafish primitive and definitive hematopoiesis. PLoS Biol. 5, e132 (2007)
Gering, M. & Patient, R. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev. Cell 8, 389–400 (2005)
Wilkinson, R. N. et al. Hedgehog signaling via a calcitonin receptor-like receptor can induce arterial differentiation independently of VEGF signaling in zebrafish. Blood 120, 477–488 (2012)
Powell, G. T. & Wright, G. J. Jamb and Jamc are essential for vertebrate myocyte fusion. PLoS Biol. 9, e1001216 (2011)
Powell, G. T. & Wright, G. J. Genomic organisation, embryonic expression and biochemical interactions of the zebrafish junctional adhesion molecule family of receptors. PLoS ONE 7, e40810 (2012)
Sakaguchi, T. et al. Putative “stemness” gene jam-B is not required for maintenance of stem cell state in embryonic, neural, or hematopoietic stem cells. Mol. Cell. Biol. 26, 6557–6570 (2006)
Arcangeli, M. L. et al. JAM-B regulates maintenance of hematopoietic stem cells in the bone marrow. Blood 118, 4609–4619 (2011)
Parsons, M. J. et al. Notch-responsive cells initiate the secondary transition in larval zebrafish pancreas. Mech. Dev. 126, 898–912 (2009)
Ahimou, F., Mok, L. P., Bardot, B. & Wesley, C. The adhesion force of Notch with Delta and the rate of Notch signaling. J. Cell Biol. 167, 1217–1229 (2004)
Westerfield, M. The Zebrafish Book: a Guide for the Laboratory Use of Zebrafish (Danio rerio) (Univ. Oregon Press, 2000)
North, T. E. et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature 447, 1007–1011 (2007)
Chi, N. C. et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734–739 (2008)
Lawson, N. D. & Weinstein, B. M. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev. Biol. 248, 307–318 (2002)
Villefranc, J. A., Amigo, J. & Lawson, N. D. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236, 3077–3087 (2007)
Distel, M., Wullimann, M. F. & Koster, R. W. Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl Acad. Sci. USA 106, 13365–13370 (2009)
Higashijima, S., Okamoto, H., Ueno, N., Hotta, Y. & Eguchi, G. High-frequency generation of transgenic zebrafish which reliably express GFP in whole muscles or the whole body by using promoters of zebrafish origin. Dev. Biol. 192, 289–299 (1997)
Zygmunt, T. et al. Semaphorin-PlexinD1 signaling limits angiogenic potential via the VEGF decoy receptor sFlt1. Dev. Cell 21, 301–314 (2011)
Kobayashi, I. et al. Characterization and localization of side population (SP) cells in zebrafish kidney hematopoietic tissue. Blood 111, 1131–1137 (2008)
Thisse, C., Thisse, B., Schilling, T. F. & Postlethwait, J. H. Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119, 1203–1215 (1993)
Fujita, N. et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol. Biol. Cell 19, 2092–2100 (2008)
Furutani-Seiki, M. et al. Neural degeneration mutants in the zebrafish, Danio rerio. Development 123, 229–239 (1996)
Acknowledgements
The authors thank G. Wright for providing the jam2ahu3319 line, A. Shimizu for help in generating transgenic lines, M. Osato for providing the I-SceI-pBSII SK+ vector, M. Distel for providing phldb1:Gal4-mCherry animals, and W. Clements, Y. Lee and E. Butko provided critical evaluation of the manuscript. This work was supported in part by a JSPS Research fellowship for Young Scientists and a JSPS Postdoctoral fellowship for Research Abroad (I.K.), by a New Investigator Award from the California Institute of Regenerative Medicine, R01-DK074482 from the National Institutes of Health, and an Innovative Science Award from the American Heart Association (D.T.).
Author information
Authors and Affiliations
Contributions
I.K., T.S. and D.T. designed research. I.K. generated transgenic lines, performed flow cytometry, cell culture and transfection experiments, analysed data, and wrote the manuscript. I.K. and J.K.-S. performed in situ hybridization and real-time PCR. I.K., J.K.-S. and C.P. generated in situ probes. I.K. and N.F. performed immunoprecipitation and western blotting. I.K. and A.D.K. performed confocal imaging. J.K.-S. performed histological analyses. A.D.K., C.P., T.S. and D.T. edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Alignment and phylogenetic analysis of Jam1a.
a, The genomic loci of the jam1a and jam1b genes. Arrows indicate the orientation of each gene. b, Alignment of zebrafish Jam1a, Jam1b, and human JAM1. The multiple alignment was produced using ClustalW. Asterisks indicate a fully conserved residue. Colons and periods indicate strong and weak similarity, respectively. Dashes indicate gaps. Boxes coloured red, signal peptide (Sp); light blue, immunoglobulin-like domain (Ig); blue, transmembrane domain (TM); green, PDZ-binding domain (PBD); yellow, coiled-coil domain (CCD). Shaded boxes show a cis-dimerization motif. c, Phylogenetic analysis of Jam proteins. Cii, Ciona intestinalis; Dre, Danio rerio; Hsa, Homo sapiens; Mmu, Mus musculus; Tni, Tetraodon nigroviridis; Gga, Gallus gallus; Ssc, Sus scrofa; Bta, Bos taurus; Fca, Felis silvestris catus. Cii Jam3 was used as an out-group. The numbers at the relevant branches refer to bootstrap values of 1,000 replications. d, Schematic diagrams of human JAM1 and zebrafish Jam1a (left) and Jam1b (right).
Extended Data Figure 2 The expression of jam1a and lineage tracing of jam1a-expressing cells.
a–c, The expression of jam1a at 14, 18 and 24 hpf. Black, blue, and red arrows indicate the posterior lateral mesoderm (PLM), pronephros, and dorsal aorta (DA), respectively. d, Single or two-colour whole-mount in situ hybridization with jam1a (purple) and/or fli1 (red) probes. The expression domain of fli1 merged with that of jam1a (arrows). e, qPCR analysis of jam1a in purified fli1:GFP+ cells at 14, 17, and 20 hpf. Relative expression levels in each cell population were calculated from the expression levels in the kidney tissue. Error bars, s.d. f, GFP expression of -2.2jam1a:GFP at 30 hpf. GFP expression was strongly detected in posterior pronephros, lateral lines, and otic vesicles. g, The expression of CreERT2 in -5.1jam1a:CreERT2 at 24 hpf. h, Three different schedules of 4-hydroxytamoxifen (4OHT) treatment (12–16 hpf, 18–22 hpf, and 24–28 hpf) in -5.1jam1a:CreERT2; bactin2:loxP-BFP-loxP-DsRed. The numbers indicate hour post fertilization (hpf), and red insets in the blue arrows indicate the period of the 4OHT treatment. i, Flow cytometric analysis of kidney marrow cells in 12–16 hpf group (left, n = 5), 18–22 hpf group (middle, n = 5), and 24–28 hpf group (right, n = 2) at 2 months of age. One 12–16 hpf animal showed switched DsRed+ cells in kidney marrow cells. j, DsRed+ cells from 12–16 hpf kidneys are distributed in all blood cell populations, including neutrophils and eosinophils (N+E), precursors and monocytes (P+M), and lymphocytes and thrombocytes (L+T). k, l, Confocal imaging of the DA and caudal haematopoietic tissue (CHT) in a 12–16 hpf embryo at 48 hpf. Right panels show high magnification views of the boxed regions in left panels. Embryos are oriented with anterior to the left. There are many ‘switched’ DsRed+ cells in the ventral floor of the DA and in the CHT (arrowheads). Bars, 20 μm. Data are representative of two independent experiments with two different clutches of embryos (a–e, g) or eight embryos (k, l) or pooled from two independent experiments (i, j).
Extended Data Figure 3 Characterization of jam1a MOex7-injected embryos.
a, RT–PCR results from jam1a MOex7-injected embryos. cDNA from embryos uninjected or injected with various doses of jam1a MOex7 (100–500 μM) was subjected to RT–PCR analysis using specific primers, which amplify from exon 4 to 10 of jam1a. ef1a was used as a control. The expected size of PCR products in uninjected embryos is 746 base pairs (bp) (black arrow). Exon 7 (108 bp)-skipped products were detected in jam1a MOex7-injected embryos (red arrow). The dose of 300 μM was used in this study. b, Exon 7-skipped products were verified by sequencing. The dotted line indicates the junctions between exon 6 and exon 7 (uninjected, upper panel) or exon 8 (jam1a MOex7, lower panel). c, A schematic diagram of the mRNA splicing in jam1a MOex7-injected embryos. The red bar indicates the binding site of jam1a MOex7. d, Schematic diagrams of Jam1a protein in wild-type (left) or jam1a MOex7-injected embryos (right). Since exon 7 encodes the transmembrane domain (TM), jam1a MOex7-injected embryos express a mutant protein lacking the TM. Sp, signal peptide; Ig, immunoglobulin-like domain; PBD, PDZ-binding domain. e, The relative expression of wild-type jam1a mRNA in uninjected or jam1a MOex7 (300 μM)-injected embryos at 24 hpf. The reverse primer was designed in exon 7. Error bars, s.d. f–i, The expression of cmyb and rag1 in uninjected or jam1a MOex7-injected embryos. Arrowheads indicate the dorsal aorta (f, g) or the thymus (h, i). Data are representative of two independent experiments with two different clutches of embryos (a, e–i).
Extended Data Figure 4 jam1a is required for HSC specification.
a–c, Fluorescently labelled HSCs in cmyb:GFP; kdrl:mCherry. The number of cmyb:GFP; kdrl:mCherry double-positive cells in the dorsal aorta (DA) were counted in uninjected or jam1a MOatg-injected embryos at 48 hpf (arrows). Embryos are oriented with anterior to the left. The average number of double-positive cells is significantly lower in jam1a MOatg-injected embryos (n = 10) compared with uninjected embryos (n = 10), *P < 0.001 by Student’s t-test; error bars, s. d. d–q, The expression of gata1 (an erythroid marker), l-plastin (a myeloid marker) at 24 hpf or 4 dpf, kdrl (a pan-endothelial marker) in the trunk or caudal haematopoietic tissue (CHT), tbx20 (a marker for the roof of DA), flt4 (a vein marker), cdh17 (a pronephros marker), desma (a somite marker), nkx3.1 (a sclerotome marker), shha (a notochord marker) at 16 hpf or 26 hpf, and vegfa at 17 hpf or 26 hpf in uninjected or jam1a MOatg-injected embryos. Arrowheads indicate CHT (f), DA (i), or notochord (o). Data are representative of two independent experiments with ten embryos (a–c) or two different clutches of embryos (d–q).
Extended Data Figure 5 Characterization of jam2a MOex5-injected embryos.
a, RT–PCR results from jam2a MOex5-injected embryos. cDNA from embryos uninjected or injected with various doses of jam2a MOex5 (200–600 μM) was subjected to RT–PCR analysis using specific primers, which amplify from exon 4 to 10 of jam2a. ef1a was used as a control. The expected size of PCR products in uninjected embryos is 537 base pairs (bp) (black arrow). Intron 4 (72 bp)-trapped products were detected in jam2a MOex5-injected embryos (red arrow). The dose of 400 μM was used in this study. b, Intron 4-trapped products were verified by sequencing. The dotted line indicates the junctions between exon 4 and exon 5 (uninjected, upper panel) or intron 4 (jam2a MOex5, lower panel). c, A schematic diagram of the mRNA splicing in jam2a MOex5-injected embryos. The red bar indicates the binding site of jam2a MOex5. d, Schematic diagrams of Jam2a protein in wild-type (left) or jam2a MOex5-injected embryos (right). Since intron 4 contains an in-frame stop codon, jam2a MOex5-injected embryos express a truncated mutant protein. Sp, signal peptide; Ig, immunoglobulin-like domain; TM, transmembrane domain; PBD, PDZ-binding domain. e–l, The expression of runx1, efnb2a, cmyb, and rag1 in uninjected or jam2a MOex5-injected embryos. Arrowheads indicate the dorsal aorta (e–j) or the thymus (k, l). m, A transverse section of a fli1:GFP; phldb1:mCherry embryo injected with jam2a MOex5 at 16 hpf. A high magnification view of the boxed region is shown in the right panel. A dotted line in the right panel indicates the contact surface area between fli1:GFP+ PLM cells and phldb1:mCherry+ somitic cells. Bars, 10 μm. n, The average contact surface area between a PLM cell and the somite in uninjected, jam1a MOatg-, or jam2a MOex5-injected embryos. The contact surface area per cell (μm per cell) was calculated from at least one hundred fli1:GFP+ cells in an embryo, and the averages were obtained from three embryos of each. *P < 0.01, by Student’s t-test. Error bars, s.d. Data are representative of two independent experiments with two different clutches of embryos (a, e–l) or three embryos (m, n).
Extended Data Figure 6 jam2a is required for HSC specification.
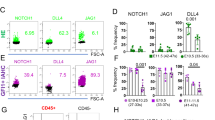
a, A schematic diagram of the preparation of jam2a mutant (jam2ahu3319) embryos. Embryos from an incross of genotyped homozygous (homo) jam2ahu3319 animals were examined by whole-mount in situ hybridization (WISH). b–g, The expression of runx1 and cmyb in the dorsal aorta (DA) and rag1 in the thymus in wild-type or jam2ahu3319 embryos. The numbers shown in each panel indicate the frequency of embryos showing each expression pattern. h, i, The expression of fli1 at 18 hpf in wild-type or jam2ahu3319 embryos. Arrows indicate a subset of fli1+ cells that did not reach the midline. j–o, The expression of endothelial marker genes (kdrl, efnb2a, and flt4) in wild-type or jam2ahu3319 embryos at 28 hpf. Approximately 90% of embryos showed normal vascular plexus, while the rest of embryos showed a reduction of efnb2a and kdrl. Arrowheads indicate the DA (b–e, l, m), the thymus (f, g), or posterior cardinal vein (n, o). p, Histological analysis of the adult kidney in a wild-type or jam2ahu3319 animal at 2 months of age. Many blood cells are observed in the marrow area. Haematoxylin and eosin (HE) staining. q, Representative results of flow cytometric analysis of kidney marrow cells from a wild-type or jam2ahu3319 animal at 3 months of age. All blood cell populations are detected in jam2ahu3319 animals. L+T, lymphocytes and thrombocytes; N+E, neutrophils and eosinophils; P+M, precursors and monocytes. Data are representative of two independent experiments with two different clutches of embryos (b–o) or three different animals (p, q).
Extended Data Figure 7 Enforced expression of the Notch intracellular domain rescues HSCs in jam1a or jam2a morphants.
Heat-shock (HS) (hsp70:Gal4, a, b, e) or endothelial (fli1:Gal4, c, d, f) induction of Notch intracellular domain (NICD) in uninjected, jam1a MOatg-, or jam2a MOatg-injected embryos. Left panels show whole-mount immunofluorescence visualization of Myc-tagged NICD, and right panels show the expression of runx1 at 26 hpf. Arrowheads indicate the dorsal aorta. Data are representative of two independent experiments with two different clutches of embryos (a–f).
Extended Data Figure 8 Somitic Dlc and Dld are involved in the activation of endothelial Notch signalling.
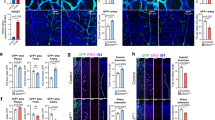
a, b, Transverse sections of fli1:GFP embryos stained with dlc or dld (purple) and anti-GFP antibody (brown) at 15 hpf. Right panels show high magnification views of the boxed regions. Migrating fli1:GFP+ cells (black arrowheads) are in contact with dlc+ or dld+ somitic cells (white arrowheads). c–e, Flow cytometric analysis of Tp1:GFP; fli1:DsRed embryos uninjected or injected with wnt16 MO at 22 hpf. Representative results of flow cytometric analysis (c), the mean fluorescent intensities of GFP in Tp1:GFP+; fli1:DsRed+ populations (d), and the percentages of Tp1:GFPhigh in fli1:DsRed+ populations (e) are shown. Blue gates and red circles indicate the Tp1:GFP+; fli1:DsRed+ and Tp1:GFPhigh; fli1:DsRed+ population, respectively. *P < 0.01, by Student’s t-test. Error bars, s.d. f, g, Lateral views of the dorsal aorta (DA) in Tp1:GFP; fli1:DsRed embryos uninjected or injected with wnt16 MO at 28 hpf. Arrows indicate the low activation of Tp1:GFP in the ventral floor of the DA. Data are representative of two independent experiments with four embryos (a, b), eight embryos (f, g), or four different clutches of embryos (c–e).
Extended Data Figure 9 Aortic Tp1:GFP expression is restored by overexpression of dlc or dld in jam1a morphants.
a–h, The aortic expression of notch1b, notch3, dlc, and dll4 in uninjected or jam1a MOatg-injected embryos at 26 hpf. Arrowheads indicate the dorsal aorta (DA). i–p, Lateral views of the DA in Tp1:GFP (i–l) and transverse sections of Tp1:GFP; fli1:DsRed (m–p) at 28 hpf. Embryos were uninjected, injected with jam1a MOatg alone, or co-injected with jam1a MOatg and dlc or dld mRNA. Arrows indicate relatively low activation of Tp1:GFP in the ventral floor of the DA. The expression of Tp1:GFP was restored in the ventral floor of the DA by co-injection with dlc or dld. Bars, 10 μm. Data are representative of two independent experiments with two different clutches of embryos (a–h), eight embryos (i–l), or three embryos (m–p).
Extended Data Figure 10 A model of Notch signal transduction in HSC precursors.
Jam1a+ PLM cells initially flank the somites then migrate to the midline along the ventral face of the somite, where Jam2a and the Notch ligands Dlc and Dld are expressed. Binding of Jam1a and Jam2a in trans is required for transmission of Notch signals into the PLM derivatives that subsequently generate aortic haemogenic endothelium (left side). In jam1a-deficient embryos, although PLM cells arise and initially migrate normally, their migration is delayed upon contact with the somite. Moreover, they show low activation of Notch signalling due to poor interaction with the somite, resulting in the failure of HSC specification in the aortic floor (right side).
Supplementary information
Supplementary Information
This file contains Supplementary Tables 1-2. (PDF 101 kb)
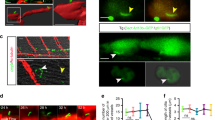
PLM cell migration in uninjected wild type embryo
A time-lapse imaging of uninjected fli1:GFP; phldb1:Gal4-mCherry double transgenic embryo. The video was captured from 14hpf onwards. The embryo is shown in a dorsal view, with head to the left. Elapsed time is shown at the top right. (MOV 9484 kb)
PLM cell migration in jam1a MOatg-injected embryo
A time-lapse imaging of fli1:GFP; phldb1:Gal4-mCherry double transgenic embryo injected with jam1a MOatg. The video was captured from 14hpf onwards. The embryo is shown in a dorsal view, with head to the left. Elapsed time is shown at the top right. (MOV 9424 kb)
PLM cell migration in jam1a MOex7-injected embryo
A time-lapse imaging of fli1:GFP; phldb1:Gal4-mCherry double transgenic embryo injected with jam1a MOex7. The video was captured from 14hpf onwards. The embryo is shown in a dorsal view, with head to the left. Elapsed time is shown at the top right. (MOV 9447 kb)
PLM cell migration in jam2a MO-injected embryo
A time-lapse imaging of fli1:GFP; phldb1:Gal4-mCherry double transgenic embryo injected with jam2a MO. The video was captured from 14hpf onwards. The embryo is shown in a dorsal view, with head to the left. Elapsed time is shown at the top right. It should be noted that jam2a morphants show torsion in the development of the left-right body axis. (MOV 9435 kb)
PLM cell migration in jam2a MOex5-injected embryo
A time-lapse imaging of fli1:GFP; phldb1:Gal4-mCherry double transgenic embryo injected with jam2a MOex5. The video was captured from 14hpf onwards. The embryo is shown in a dorsal view, with head to the left. Elapsed time is shown at the top right. (MOV 9282 kb)
Rights and permissions
About this article
Cite this article
Kobayashi, I., Kobayashi-Sun, J., Kim, A. et al. Jam1a–Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature 512, 319–323 (2014). https://doi.org/10.1038/nature13623
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature13623
This article is cited by
-
Glia maturation factor-γ is required for initiation and maintenance of hematopoietic stem and progenitor cells
Stem Cell Research & Therapy (2023)
-
Alcam-a and Pdgfr-α are essential for the development of sclerotome-derived stromal cells that support hematopoiesis
Nature Communications (2023)
-
Ligament injury in adult zebrafish triggers ECM remodeling and cell dedifferentiation for scar-free regeneration
npj Regenerative Medicine (2023)
-
Lifelong single-cell profiling of cranial neural crest diversification in zebrafish
Nature Communications (2022)
-
The F11 Receptor (F11R)/Junctional Adhesion Molecule-A (JAM-A) (F11R/JAM-A) in cancer progression
Molecular and Cellular Biochemistry (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.